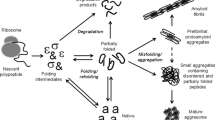
Abstract
Aggresomes are protein aggregates found in mammalian cells when the intracellular protein degradation machinery is over-titered. Despite that they abound in cells producing recombinant proteins of biomedical and biotechnological interest, the physiological roles of these protein clusters and the functional status of the embedded proteins remain basically unexplored. In this work, we have determined for the first time that, like in bacterial inclusion bodies, deposition of recombinant proteins into aggresomes does not imply functional inactivation. As a model, human α-galactosidase A (GLA) has been expressed in mammalian cells as enzymatically active, mechanically stable aggresomes showing higher thermal stability than the soluble GLA version. Since aggresomes are easily produced and purified, we propose these particles as novel functional biomaterials with potential as carrier-free, self-immobilized catalyzers in biotechnology and biomedicine.





Similar content being viewed by others
References
Berrow NS, Alderton D, Sainsbury S, Nettleship J, Assenberg R, Rahman N, Stuart DI, Owens RJ (2007) A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res 35, e45
Cano-Garrido O, Rodriguez-Carmona E, ez-Gil C, Vazquez E, Elizondo E, Cubarsi R, Seras-Franzoso J, Corchero JL, Rinas U, Ratera I, Ventosa N, Veciana J, Villaverde A, Garcia-Fruitos E (2013) Supramolecular organization of protein-releasing functional amyloids solved in bacterial inclusion bodies. Acta Biomater 9:6134–6142
Chiba Y, Takei S, Kawamura N, Kawaguchi Y, Sasaki K, Hasegawa-Ishii S, Furukawa A, Hosokawa M, Shimada A (2012) Immunohistochemical localization of aggresomal proteins in glial cytoplasmic inclusions in multiple system atrophy. Neuropathol Appl Neurobiol 38:559–571
Corchero JL, Mendoza R, Lorenzo J, Rodriguez-Sureda V, Dominguez C, Vazquez E, Ferrer-Miralles N, Villaverde A (2011) Integrated approach to produce a recombinant, his-tagged human alpha-galactosidase a in mammalian cells. Biotechnol Prog 27:1206–1217
Desnick RJ, Allen KY, Desnick SJ, Raman MK, Bernlohr RW, Krivit W (1973) Fabry's disease: enzymatic diagnosis of hemizygotes and heterozygotes. Alpha-galactosidase activities in plasma, serum, urine, and leukocytes. J Lab Clin Med 81:157–171
Garcia-Fruitos E, Gonzalez-Montalban N, Morell M, Vera A, Ferraz RM, Aris A, Ventura S, Villaverde A (2005) Aggregation as bacterial inclusion bodies does not imply inactivation of enzymes and fluorescent proteins. Microb Cell Factories 4:27
Garcia-Fruitos E, Aris A, Villaverde A (2007) Localization of functional polypeptides in bacterial inclusion bodies. Appl Environ Microbiol 73:289–294
Garcia-Mata R, Bebok Z, Sorscher EJ, Sztul ES (1999) Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J Cell Biol 146:1239–1254
Garcia-Mata R, Gao YS, Sztul E (2002) Hassles with taking out the garbage: aggravating aggresomes. Traffic 3:388–396
Gershon ND, Porter KR, Trus BL (1985) The cytoplasmic matrix: its volume and surface area and the diffusion of molecules through it. Proc Natl Acad Sci U S A 82:5030–5034
Ioannou YA, Bishop DF, Desnick RJ (1992) Overexpression of human alpha-galactosidase A results in its intracellular aggregation, crystallization in lysosomes, and selective secretion. J Cell Biol 119:1137–1150
Ioannou YA, Zeidner KM, Grace ME, Desnick RJ (1998) Human alpha-galactosidase A: glycosylation site 3 is essential for enzyme solubility. Biochem J 332(Pt 3):789–797
Johnston JA, Ward CL, Kopito RR (1998) Aggresomes: a cellular response to misfolded proteins. J Cell Biol 143:1883–1898
Johnston JA, Dalton MJ, Gurney ME, Kopito RR (2000) Formation of high molecular weight complexes of mutant Cu, Zn-superoxide dismutase in a mouse model for familial amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A 97:12571–12576
Johnston JA, Illing ME, Kopito RR (2002) Cytoplasmic dynein/dynactin mediates the assembly of aggresomes. Cell Motil Cytoskeleton 53:26–38
Junn E, Lee SS, Suhr UT, Mouradian MM (2002) Parkin accumulation in aggresomes due to proteasome impairment. J Biol Chem 277:47870–47877
Kopito RR (2000) Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol 10:524–530
Kopito RR, Sitia R (2000) Aggresomes and Russell bodies. Symptoms of cellular indigestion? EMBO Rep 1:225–231
Mayes JS, Scheerer JB, Sifers RN, Donaldson ML (1981) Differential assay for lysosomal alpha-galactosidases in human tissues and its application to Fabry's disease. Clin Chim Acta 112:247–251
Nahalka J (2008) Physiological aggregation of maltodextrin phosphorylase from Pyrococcus furiosus and its application in a process of batch starch degradation to alpha-D-glucose-1-phosphate. J Ind Microbiol Biotechnol 35:219–223
Nahalka J, Nidetzky B (2007) Fusion to a pull-down domain: a novel approach of producing Trigonopsis variabilis D-amino acid oxidase as insoluble enzyme aggregates. Biotechnol Bioeng 97:454–461
Nahalka J, Patoprsty V (2009) Enzymatic synthesis of sialylation substrates powered by a novel polyphosphate kinase (PPK3). Org Biomol Chem 7:1778–1780
Nahalka J, Gemeiner P, Bucko M, Wang PG (2006) Bioenergy beads: a tool for regeneration of ATP/NTP in biocatalytic synthesis. Artif Cells Blood Substit Immobil Biotechnol 34:515–521
Nahalka J, Dib I, Nidetzky B (2008a) Encapsulation of Trigonopsis variabilis D-amino acid oxidase and fast comparison of the operational stabilities of free and immobilized preparations of the enzyme. Biotechnol Bioeng 99:251–260
Nahalka J, Vikartovska A, Hrabarova E (2008b) A crosslinked inclusion body process for sialic acid synthesis. J Biotechnol 134:146–153
Seras-Franzoso J, Peebo K, Corchero JL, Tsimbouri PM, Unzueta U, Rinas U, Dalby MJ, Vazquez E, Garcia-Fruitos E, Villaverde A (2013a) A nanostructured bacterial bioscaffold for the sustained bottom-up delivery of protein drugs. Nanomedicine (Lond)
Seras-Franzoso J, Steurer C, Roldan M, Vendrell M, Vidaurre-Agut C, Tarruella A, Saldana L, Vilaboa N, Parera M, Elizondo E, Ratera I, Ventosa N, Veciana J, Campillo-Fernandez AJ, Garcia-Fruitos E, Vazquez E, Villaverde A (2013b) Functionalization of 3D scaffolds with protein-releasing biomaterials for intracellular delivery. J Control Release 171:63–72
Seras-Franzoso J, Peebo K, Garcia-Fruitos E, Vazquez E, Rinas U, Villaverde A (2014) Improving protein delivery of fibroblast growth factor-2 from bacterial inclusion bodies used as cell culture substrates. Acta Biomater 10:1354–1359
Shimohata T, Sato A, Burke JR, Strittmatter WJ, Tsuji S, Onodera O (2002) Expanded polyglutamine stretches form an 'aggresome'. Neurosci Lett 323:215–218
Tokatlidis K, Dhurjati P, Millet J, Beguin P, Aubert JP (1991) High activity of inclusion bodies formed in Escherichia coli overproducing Clostridium thermocellum endoglucanase D. FEBS Lett 282:205–208
Upadhyay AK, Murmu A, Singh A, Panda AK (2012) Kinetics of inclusion body formation and its correlation with the characteristics of protein aggregates in Escherichia coli. PLoS One 7, e33951
Vazquez E, Corchero JL, Burgueno JF, Seras-Franzoso J, Kosoy A, Bosser R, Mendoza R, Martinez-Lainez JM, Rinas U, Fernandez E, Ruiz-Avila L, Garcia-Fruitos E, Villaverde A (2012) Functional inclusion bodies produced in bacteria as naturally occurring nanopills for advanced cell therapies. Adv Mater 24:1742–1747
Wang Y, Meriin AB, Costello CE, Sherman MY (2007) Characterization of proteins associated with polyglutamine aggregates: a novel approach towards isolation of aggregates from protein conformation disorders. Prion 1:128–135
Wigley WC, Fabunmi RP, Lee MG, Marino CR, Muallem S, DeMartino GN, Thomas PJ (1999) Dynamic association of proteasomal machinery with the centrosome. J Cell Biol 145:481–490
Worrall DM, Goss NH (1989) The formation of biologically active beta-galactosidase inclusion bodies in Escherichia coli. Aust J Biotechnol 3:28–32
Zhang X, Qian SB (2011) Chaperone-mediated hierarchical control in targeting misfolded proteins to aggresomes. Mol Biol Cell 22:3277–3288
Acknowledgments
MALDI-TOF analyses were carried out in the Proteomics and Structural Biology Facility (SepBioEs) from UAB, member of ProteoRed network. We thank the Protein Expression Core Facility, Institute for Research in Biomedicine (IRB, Barcelona) for cloning the GLA gene into the pOPIN vector. We appreciate the technical support from the UAB Scientific and Technical Services SCAC (Servei de Cultius Cel · lulars, Producció d'Anticossos i Citometria). Protein production has been partially performed by the ICTS “NANBIOSIS,” more specifically by the Protein Production Platform of CIBER in Bioengineering, Biomaterials & Nanomedicine (CIBER-BBN)/IBB, at the UAB (http://www.ciber-bbn.es/en/programas/89-plataforma-de-produccion-de-proteinas-ppp). Authors appreciate the financial support through the grant EUI2008-03610 (MICINN) linked to the ERANET-IB 08–007 project, from Agència de Gestió d’Ajuts Universitaris i de Recerca (2014SGR-132), and for the grant to the project “Development of nanomedicinas for enzymatic replacement therapy in Fabry disease,” granted by the Fundació Marató TV3. We also acknowledge the support of the CIBER de Bioingeniería, Biomateriales y Nanomedicina (CIBER-BBN), an initiative funded by the VI National R&D&i Plan 2008–2011, Iniciativa Ingenio 2010, Consolider Program, CIBER Actions and financed by the Instituto de Salud Carlos III with assistance from the European Regional Development Fund. AV was distinguished with an ICREA ACADEMIA award (Catalonia, Spain).
Conflict of interest
The authors declare no commercial or financial conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rodríguez-Carmona, E., Mendoza, R., Ruiz-Cánovas, E. et al. A novel bio-functional material based on mammalian cell aggresomes. Appl Microbiol Biotechnol 99, 7079–7088 (2015). https://doi.org/10.1007/s00253-015-6684-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6684-0