Abstract
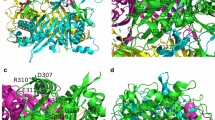
Bacillus fastidious uricase (BF uricase) containing 322 amino acid residues exhibited high stability under physiological conditions. Its crystal structure was solved to 1.4-Å resolution, showing homotetramer containing two homodimers. After the intersubunit antiparallel β-sheet in its homodimer, each subunit had a total of 18 C-terminus residues forming an α-helix (Q305-A313) and random coil (S314-L322) on surface to bury other two α-helices (I227-T238 and I244-R258). In comparison, reported crystal structures of Arthrobacter globiformis and Aspergillus flavus uricases had atomic coordinates of only some C-terminus residues, while the crystal structures of all the other uricases accessible before September 2014 missed atomic coordinates of all their C-terminus residues, after the intersubunit antiparallel β-sheets. In each homodimer of BF uricase, H-bonds were found between E311 and Y249 and between Y319 and D257; electrostatic interaction networks were found to surround D307 plus R310 and intersubunit R3, K312 plus D257, E318 plus K242, and L322 plus R258. Amino acid mutations that disrupted those interactions when R3 and D307 were reserved caused moderate decreases of activity at pH 9.2 while negligible decreases of activity at pH 7.4, but destroyed stability at pH 7.4 while slightly decreased stability at pH 9.2. Such structural information guided the fusion of 6His-tag to the C-terminus of the mutant L322D with SNSNSN as a linker to reserve the activity and stability. Hence, the folding of the C-terminus residues is crucial for thermal stability of BF uricase under physiological conditions; these new structural insights are valuable for molecular engineering of uricases.




Similar content being viewed by others
References
Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221
Alvares K, Widrow RJ, Abu-Jawdeh GM, Schmidt JV, Yeldandi AV, Rao MS, Reddy JK (1992) Rat urate oxidase produced by recombinant baculovirus expression: formation of peroxisome crystalloid core-like structures. Proc Natl Acad Sci U S A 89:4908–4912
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Chan CH, Wilbanks CC, Makhatadze GI, Wong KB (2012) Electrostatic contribution of surface charge residues to the stability of a thermophilic protein: benchmarking experimental and predicted pKa values. PLoS One 7(1):e30296
Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21
Chica RA, Doucet N, Pelletier JN (2005) Semi-rational approaches to engineering enzyme activity: combining the benefits of directed evolution and rational design. Curr Opin Biotechnol 16:378–384
Colloc’h N, El Hajji M, Bachet B, L’Hermite G, Schiltz M, Prangé T, Castro B, Mornon JP (1997) Crystal structure of the protein drug urate oxidase-inhibitor complex at 2.05 Å resolution. Nat Struct Biol 4:947–952
Colloc’h N, Gabison L, Monard G, Altarsha M, Chiadmi M, Marassio G, Sopkova-de Oliveira Santos J, El Hajji M, Castro B, Abraini JH, Prangé T (2008) Oxygen pressurized X-ray crystallography: probing the dioxygen binding site in cofactorless urate oxidase and implications for its catalytic mechanism. Biophys J 95:2415–2422
Crittenden DB, Pillinger MH (2013) New therapies for gout. Annu Rev Med 64:325–337
Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB 3rd, Snoeyink J, Richardson JS, Richardson DC (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35:W375–W383
Der BS, Kluwe C, Miklos AE, Jacak R, Lyskov S, Gray JJ, Georgiou G, Ellington AD, Kuhlman B (2013) Alternative computational protocols for supercharging protein surfaces for reversible unfolding and retention of stability. PLoS One 8(5):e64363
Ea HK, Chales G, Lioté F (2011) Pegloticase and chronic gout. JAMA 306:1979
Feng J, Li X, Yang X, Zhang C, Yuan Y, Pu J, Zhao Y, Xie Y, Yuan H, Bu Y, Liao F (2010) A new practical system for evaluating the pharmacological properties of uricase as a potential drug for hyperuricemia. Arch Pharm Res 33:1761–1769
Feng J, Liu H, Yang X, Gao A, Liao J, Feng L, Pu J, Xie Y, Long G, Li Y, Liao F (2013) Comparison of activity indexes for recognizing enzyme mutants of higher activity with uricase as model. Chem Cent J 7:69
Gabison L, Prangé T, El Colloc’h N, Hajji M, Castro B, Chiadmi M (2008) Structural analysis of urate oxidase in complex with its natural substrate inhibited by cyanide: mechanistic implications. BMC Struct Biol 8:32
Gabison L, Chiadmi M, El Hajji M, Castro B, Colloc’h N, Prangé T (2010) Near-atomic resolution structures of urate oxidase complexed with its substrate and analogues: the protonation state of the ligand. Acta Crystallogr D Biol Crystallogr 66:714–724
Hibi T, Hayashi Y, Fukada H, Itoh T, Nago T, Nishiya Y (2014) Intersubunit salt bridges with a sulfate anion control subunit dissociation and thermal stabilization of Bacillus sp. TB-90 urate oxidase. Biochemistry 53:3879–3888
Huang Y, Chen Y, Yang X, Zhao H, Hu X, Pu J, Liao J, Long G, Liao F (2015) Optimization of pH values to formulate the bireagent kit for serum uric acid assay. Biotechnol Appl Biochem 62:137–144
Juan EC, Hoque MM, Shimizu S, Hossain MT, Yamamoto T, Imamura S, Suzuki K, Tsunoda M, Amano H, Sekiguchi T, Takénaka A (2008) Structures of Arthrobacter globiformis urate oxidase-ligand complexes. Acta Crystallogr D Biol Crystallogr D64:815–822
Kahn K, Tipton PA (1997) Kinetic mechanism and cofactor content of soybean root nodule urate oxidase. Biochemistry 36:4731–4738
Li Y, Yang X, He C, Hu X, Pu J, Liu L, Long G, Liao F (2014) Facile quantitative comparison of specific activities of fusion-tagged enzyme/mutants in cell lysates via prediction of their maximum adsorption by anti-tag antibody immobilized in microplate wells. RSC Adv 4:29925–29932
Li Y, Long G, Yang X, Hu X, Feng Y, Tan D, Xie Y, Pu J, Liao F (2015) Approximated maximum adsorption of His-tagged enzyme/mutants on Ni2+-NTA for comparison of specific activities. Int J Biol Macromol 74C:211–217
Liao F, Zhu XY, Wang YM, Zuo YP (2005) The comparison of the estimation of enzyme kinetic parameters by fitting reaction curve to the integrated Michaelis-Menten rate equations of different predictor variables. J Biochem Biophys Methods 62:13–24
Liao F, Zhao YS, Zhao LN, Tao J, Zhu XY, Liu L (2006) Evaluation of a kinetic uricase method for serum uric acid assay by predicting background absorbance of uricase reaction solution with an integrated method. J Zhejiang Univ Sci B 7:497–502
Liu Z, Lu D, Li J, Chen W, Liu Z (2009) Strengthening intersubunit hydrogen bonds for enhanced stability of recombinant urate oxidase from Aspergillus flavus: molecular simulations and experimental validation. Phys Chem Chem Phys 11:333–340
Liu X, Wen M, Li J, Zhai F, Ruan J, Zhang L, Li S (2011) High-yield expression, purification, characterization and structure determination of tag-free Candida utilis uricase. Appl Microbiol Biotechnol 92:529–537
McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40:658–674
Oksanen E, Blakeley MP, El-Hajji M, Ryde U, Budayova-Spano M (2014) The neutron structure of urate oxidase resolves a long-standing mechanistic conundrum and reveals unexpected changes in protonation. PLoS One 9:e86651
Pfrimer P, de Moraes LM, Galdino AS, Salles LP, Reis VC, De Marco JL, Prates MV Jr, Bloch C, Torres FA (2010) Cloning, purification, and partial characterization of Bacillus subtilis urate oxidase expressed in Escherichia coli. J Biomed Biotechnol 2010:674908. doi:10.1155/2010/674908
Raghunathan G, Sokalingam S, Soundrarajan N, Madan B, Munussami G, Lee SG (2013) Modulation of protein stability and aggregation properties by surface charge engineering. Mol Biosyst 9(9):2379–2389
Retailleau P, Colloc’h N, Vivarès D, Bonneté F, Castro B, El-Hajji M, Mornon JP, Monard G, Prangé T (2004) Complexed and ligand-free high-resolution structures of urate oxidase (Uox) from Aspergillus flavus: a reassignment of the active-site binding mode. Acta Crystallogr D Biol Crystallogr 60:453–462
Retailleau P, Colloc’h N, Vivarès D, Bonneté F, Castro B, El Hajji M, Prangé T (2005) Urate oxidase from Aspergillus flavus: new crystal-packing contacts in relation to the content of the active site. Acta Crystallogr D Biol Crystallogr 61:218–229
Sarkissian CN, Gámez A, Wang L, Charbonneau M, Fitzpatrick P, Lemontt JF, Zhao B, Vellard M, Bell SM, Henschell C, Lambert A, Tsuruda L, Stevens RC, Scriver CR (2008) Preclinical evaluation of multiple species of PEGylated recombinant phenylalanine ammonia lyase for the treatment of phenylketonuria. Proc Natl Acad Sci U S A 105:20894–20899
Schiavon O, Caliceti P, Ferruti P, Veronese FM (2000) Therapeutic proteins: a comparison of chemical and biological properties of uricase conjugated to linear or branched poly(ethylene glycol) and poly(N-acryloylmorpholine). Farmaco 55:264–269
Schlesinger N, Yasothan U, Kirkpatrick P (2011) Pegloticase. Nat Rev Drug Discov 10:17–18
Suzuki K, Sakasegawa S, Misaki H, Sugiyama M (2004) Molecular cloning and expression of uricase gene from Arthrobacter globiformis in Escherichia coli and characterization of the gene product. J Biosci Bioeng 98:153–158
Tan Q, Zhang J, Wang N, Li X, Xiong H, Teng Y, He D, Wu J, Zhao C, Yin H, Zhang L (2012) Uricase from Bacillus fastidious loaded in alkaline enzymosomes: enhanced biochemical and pharmacological characteristics in hypouricemic rats. Eur J Pharm Biopharm 82:43–48
Turner NJ (2009) Directed evolution drives the next generation of biocatalysts. Nat Chem Biol 5:567–573
Wu S, Chen B, Liu C, Ou Y, Yi J (2009) Expression in Escherichia coli, purification and enzymatic properties of porcine urate oxidase. Sheng Wu Gong Cheng Xue Bao 25:1664–1670
Yamamoto K, Kojima Y, Kikuchi T, Shigyo T, Sugihara K, Takashio M, Emi S (1996) Nucleotide sequence of the uricase gene from Bacillus sp.TB-90. J Biochem 119:80–84
Yang X, Yuan Y, Zhan CG, Liao F (2012) Uricases as therapeutic agents to treat refractory gout: current states and future directions. Drug Dev Res 73:66–72
Zhang C, Yang X, Feng J, Yuan Y, Li X, Bu Y, Xie Y, Yuan H, Liao F (2010) Effects of modification of amino groups with poly(ethylene glycol) on a recombinant uricase from Bacillus fastidiosus. Biosci Biotechnol Biochem 74:1298–1301
Zhang P, Xu L, Li Q, Lin X, Liu H, Ma X (2012) Cloning and characterization of a thermostable urate oxidase from Microbacterium sp. strain ZZJ4-1. Sheng Wu Gong Cheng Xue Bao 28:813–822
Zhang C, Yang X, Gao A, Hu X, Pu J, Liu H, Feng J, Liao J, Li Y, Liao F (2014) Comparison of modification of a bacterial uricase with N-hydroxysuccinimide esters of succinate and carbonate of monomethoxyl poly(ethylene glycol). Biotechnol Appl Biochem 61:683-690
Zhao Y, Zhao L, Yang G, Tao J, Bu Y, Liao F (2006) Characterization of a uricase from Bacillus fastidious ATCC26904 and its application to serum uric acid assay by a patented kinetic uricase method. Biotechnol Appl Biochem 45:75–80
Zhao Y, Yang X, Lu W, Liao H, Liao F (2009a) Uricase based method for determination of uric acid in serum. Microchim Acta 164:1–6
Zhao Y, Yang X, Li X, Bu Y, Deng P, Zhang C, Feng J, Xie Y, Zhu S, Yuan H, Yu M, Liao F (2009b) Reversible inactivation of an intracellular uricase from Bacillus fastidiosus via dissociation of homotetramer into homodimers in solutions of low ionic strength. Biosci Biotechnol Biochem 73:2141–2144
Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC, Adams PD (2008) Automated structure solution with the PHENIX suite. Methods Mol Biol 426:419–435
Acknowledgments
This work was supported by the National Natural Science Foundation of China (no.30672139), Natural Sciences Foundation of CQ (nos. CSTC2011BA5039 and CSTC2012JJA0057), the Education Ministry of China (grant no.20125503110007), and the National Science Foundation of USA (grant CHE-1111761). We thank scientists in Shanghai Synchrotron Radiation Facility for technical support during collection of data.
Author information
Authors and Affiliations
Corresponding author
Additional information
Juan Feng, Lu Wang and Hongbo Liu contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 3360 kb)
Rights and permissions
About this article
Cite this article
Feng, J., Wang, L., Liu, H. et al. Crystal structure of Bacillus fastidious uricase reveals an unexpected folding of the C-terminus residues crucial for thermostability under physiological conditions. Appl Microbiol Biotechnol 99, 7973–7986 (2015). https://doi.org/10.1007/s00253-015-6520-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6520-6