Abstract
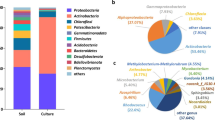
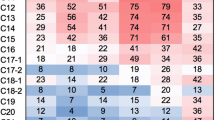
A bacterial consortium (Mix3) composed of microorganisms originating from different environments (soils and wastewater) was obtained after enrichment in the presence of a mixture of 16 hydrocarbons, gasoline, and diesel oil additives. After addition of the mixture, the development of the microbial composition of Mix3 was monitored at three different times (35, 113, and 222 days) using fingerprinting method and dominant bacterial species were identified. In parallel, 14 bacteria were isolated after 113 days and identified. Degradation capacities for Mix3 and the isolated bacterial strains were characterized and compared. At day 113, we induced the expression of catabolic genes in Mix3 by adding the substrate mixture to resting cells and the metatranscriptome was analyzed. After addition of the substrate mixture, the relative abundance of Actinobacteria increased at day 222 while a shift between Rhodococcus and Mycobacterium was observed after 113 days. Mix3 was able to degrade 13 compounds completely, with partial degradation of isooctane and 2-ethylhexyl nitrate, but tert-butyl alcohol was not degraded. Rhodococcus wratislaviensis strain IFP 2016 isolated from Mix3 showed almost the same degradation capacities as Mix3: these results were not observed with the other isolated strains. Transcriptomic results revealed that Actinobacteria and in particular, Rhodococcus species, were major contributors in terms of total and catabolic gene transcripts while other species were involved in cyclohexane degradation. Not all the microorganisms identified at day 113 were active except R. wratislaviensis IFP 2016 that appeared to be a major player in the degradation activity observed in Mix3.


Similar content being viewed by others
References
Auffret M, Labbé D, Thouand G, Greer CW, Fayolle-Guichard F (2009) Degradation of a mixture of hydrocarbons, gasoline, and diesel oil additives by Rhodococcus aetherivorans and Rhodococcus wratislaviensis. Appl Environ Microbiol 75(24):7774–7782
Baldwin BR, Mesarch MB, Nies L (2000) Broad substrate specificity of naphthalene- and biphenyl-utilizing bacteria. Appl Microbiol Biotechnol 53:748–753
Bell TH, Yergeau E, Maynard C, Juck D, Whyte LG, Greer CW (2013) Predictable bacterial composition and hydrocarbon degradation in Arctic soils following diesel and nutrient disturbance. ISME J 7:1200–1210
Bouchez-Naitali M, Vandecasteele JP (2008) Biosurfactants, an help in the biodegradation of hexadecane? The case of Rhodococcus and Pseudomonas strains. World J Microbiol Biotechnol 24:1901–1907
Brzostowicz PC, Walters DM, Jackson RE, Halsey KH, Ni H, Rouvière PE (2005) Proposed involvement of a soluble methane monooxygenase homologue in the cyclohexane-dependent growth of a new Brachymonas species. Environ Microbiol 7(2):179–190
Chaillan F, Le Flèche A, Bury E, Phantavong YH, Grimont P, Saliot A, Oudot J (2004) Identification and biodegradation potential of tropical aerobic hydrocarbon degrading microorganisms. Res Microbiol 155(7):587–595
Chandran P, Das N (2011) Degradation of diesel oil by immobilized Candida tropicalis and biofilm formed on gravels. Biodegradation 22(6):1181–1189
Chauvaux S, Chevalier F, Le Dantec C, Fayolle F, Miras I, Kunst F, Béguin P (2001) Cloning of a genetically unstable cytochrome P-450 gene cluster involved in degradation of the pollutant ethyl tert-butyl ether by Rhodococcus ruber. J Bacteriol 183:6551–6557
Ciric L, Philp JC, Whiteley AS (2010) Hydrocarbon utilization within a diesel-degrading bacterial consortium. FEMS Microbiol Lett 303(2):116–122
Damon C, Lehembre F, Oger-Desfeux C, Luis P, Ranger J, Fraissinet-Tachet L, Marmeisse R (2012) Metatranscriptomics reveals the diversity of genes expressed by eukaryotes in forest soils. PLoS One 7(1):e28967
Daugulis AJ, McCracken CM (2003) Microbial degradation of high and low molecular weight polyaromatic hydrocarbons in a two-phase partitioning bioreactor by two strains of Sphingomonas sp. Biotechnol Lett 25(17):1441–1444
de Menezes A, Clipson N, Doyle E (2012) Comparative metatranscriptomics reveals widespread community responses during phenanthrene degradation in soil. Environ Microbiol 14(9):2577–2588
Dejonghe W, Boon N, Seghers D, Top EM, Verstraete W (2001) Bioaugmentation of soils by increasing microbial richness: missing links. Environ Microbiol 3(10):649–657
DeRito CM, Pumphrey GM, Madsen EL (2005) Use of fieldbased stable isotope probing to identify adapted populations and track carbon flow through a phenol-degrading soil microbial community. Appl Environ Microbiol 71:7858–7865
Eaton RW, Chapman PJ (1992) Bacterial metabolism of naphthalene: construction and use of recombinant bacteria to study ring cleavage of 1,2-dihydroxynaphthalene and subsequent reactions. J Bacteriol 174(23):7542–7554
Fayolle-Guichard F, Durand J, Cheucle M, Rosell M, Michelland RJ, Tracol JP, Le Roux F, Grundman G, Atteia O, Richnow HH, Dumestre A, Benoit Y (2012) Study of an aquifer contaminated by ethyl tert-butyl ether (ETBE): site characterization and on-site bioremediation. J Hazard Mater 30:210–243
Fortin NY, Morales M, Nakagawa Y, Focht DD, Deshusses MA (2001) Methyl tert-butyl ether (MTBE) degradation by a microbial consortium. Environ Microbiol 3(6):407–416
Fortin N, Beaumier D, Lee K, Greer CW (2004) Soil washing improves the recovery of total community DNA from polluted and high organic content sediments. J Microbiol Methods 56:181–191
François A, Mathis H, Godefroy D, Piveteau P, Fayolle F, Monot F (2002) Biodegradation of methyl tert-butyl ether and other fuel oxygenates by a new strain, Mycobacterium austroafricanum IFP 2012. Appl Environ Microbiol 68(6):2754–2762
Fromin N, Hamelin J, Tarnawski S, Roesti D, Jourdain-Miserez K, Forestier N, Teyssier-Cuvelle S, Gillet F, Aragno M, Rossi P (2002) Statistical analysis of denaturing gel electrophoresis (DGE) fingerprinting patterns. Environ. Microbiol 4(11):634–643
Greer CW, Whyte LG, Niederberger TD (2010) Microbial communities in hydrocarbon-contaminated temperate, tropical, alpine, and polar soils. In: Timmis KN (ed) Handbook of Hydrocarbon and Lipid Microbiology. Springer, Berlin, pp 2313–2328
Hamamura N, Olson SH, Ward DM, Insskeep WP (2006) Microbial population dynamics associated with crude-oil biodegradation in diverse soils. Appl Environ Microbiol 72(9):63316–66324
Hendrickx B, Dejonghe W, Faber F, Boënne W, Bastiaens L, Verstraete W, Top EM, Springael D (2005) PCR-DGGE method to assess the diversity of BTEX mono-oxygenase genes at contaminated sites. FEMS Microbiol Ecol 55:2262–2273
Hesselsoe M, Bjerring ML, Henriksen K, Loll P, Nielsen JL (2008) Method for measuring substrate preferences by individual members of microbial consortia proposed for bioaugmentation. Biodegradation 19:621–633
Jechalke S, Rosell M, Martínez-Lavanchy PM, Pérez-Leiva P, Rohwerder T, Vogt C, Richnow HH (2011) Linking low-level stable isotope fractionation to expression of the cytochrome P450 monooxygenase-encoding ethB gene for elucidation of methyl tert-butyl ether biodegradation in aerated treatment pond systems. Appl Environ Microbiol 77(3):1086–1096
Jiménez N, Viñas M, Guiu-Aragonés C, BJ M, Albaigés J, Solanas AM (2011) Polyphasic approach for assessing changes in an autochthonous marine bacterial community in the presence of Prestige fuel oil and its biodegradation potential. Appl Microbiol Biotechnol 91(3):823–834
Jones DM, Douglas AG, Parkes RJ, Taylor J, Giger W, Schaffner C (1983) The recognition of biodegraded petroleum-derived aromatic hydrocarbons in recent marine sediments. Mar Pollut Bull 14(3):103–108
Kaplan CW, Kitts CL (2004) Bacterial succession in a petroleum land treatment unit. Appl Environ Microbiol 70(3):1777–1786
Korotkevych O, Josefiova J, Praveckova M, Cajthaml T, Stavelova M, Brennerova MV (2011) Functional adaptation of microbial communities from jet fuel-contaminated soil under bioremediation treatment: simulation of pollutant rebound. FEMS Microbiol Ecol 78(1):137–149
Le Digabel Y, Demanèche S, Benoit Y, Vogel TM, Fayolle-Guichard F (2013) Ethyl tert-butyl ether (ETBE) biodegradation by a syntrophic association of Rhodococcus sp. IFP 2042 and Bradyrhizobium sp. IFP 2049 isolated from a polluted aquifer. Appl Environ Microbiol 97(24):10531–10539
Lee EH, Cho KS (2008) Characterization of cyclohexane and hexane degradation by Rhodococcus sp. EC1. Chemosphere 71(9):1738–1744
Lee EH, Cho KS (2009) Effect of substrate interaction on the degradation of methyl tert-butyl ether, benzene, toluene, ethylbenzene, and xylene by Rhodococcus sp. J Hazard Mater 167:669–674
Lopes Ferreira N, Mathis H, Labbé D, Monot F, Greer CW, Fayolle-Guichard F (2007) n-Alkane assimilation and tert-butyl alcohol (TBA) oxidation capacity in Mycobacterium austroafricanum strains. Appl Microbiol Biotechnol 75(4):909–919
Lopes-Ferreira N, Labbé D, Monot F, Fayolle-Guichard F, Greer CW (2006) Genes involved in the methyl tert-butyl ether (MTBE) metabolic pathway of Mycobacterium austroafricanum IFP 2012. Microbiology 152:1361–1374
Ma YF, Wang L, Shao ZZ (2006) Pseudomonas, the dominant polycyclic aromatic hydrocarbon-degrading bacteria isolated from Antarctic soils and the role of large plasmids in horizontal gene transfer. Environ Microbiol 8:455–465
Malandain C, Fayolle-Guichard F, Vogel TM (2010) Cytochromes P450-mediated degradation of fuel oxygenates by environmental isolates. FEMS Microbiol Ecol 72(2):289–296
Mariano AP, Bonotto DM, de Angelis DD, Pirollo MPS, Contiero J (2008) Biodegradability of commercial and weathered diesel oils. Braz J Microbiol 39:133–142
Martienssen M, Fabritius H, Kukla S, Balcke GU, Hasselwander E, Schirmer M (2006) Determination of naturally occurring MTBE biodegradation by analysing metabolites and biodegradation by-products. J Contam Hydrol 87(1–2):37–53
Marzorati M, Wittebolle L, Boon N, Daffonchio D, and Verstraete W (2008) How to get more out of molecular fingerprints: practical tools for microbial ecology. Environ Microbiol 10(6):1571–1581
Mason OU, Hazen TC, Borglin S, Chain PS, Dubinsky EA, Fortney JL, Han J, Holman HY, Hultman J, Lamendella R, Mackelprang R, Malfatti S, Tom LM, Tringe SG, Woyke T, Zhou J, Rubin EM, Jansson JK (2012) Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to Deepwater Horizon oil spill. ISME J 6(9):1715–1727
McKay DB, Prucha M, Reineke W, Timmis KN, Pieper DH (2003) Substrate specificity and expression of three 2,3-dihydroxybiphenyl 1,2-dioxygenases from Rhodococcus globerulus strain P6. J Bacteriol 185(9):2944–2951
Medina-Bellver JI, Marín P, Delgado A, Rodríguez-Sánchez A, Reyes E, Ramos JL, Marqués S (2005) Evidence for in situ crude oil biodegradation after the Prestige oil spill. Environ Microbiol 7(6):773–779
Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA (2008) The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinforma 9:386
Müller RH, Rohwerder T, Harms H (2008) Degradation of fuel oxygenates and their main intermediates by Aquincola tertiaricarbonis L108. Microbiology 154(5):1414–1421
Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59(3):695–700
Nicolau E, Kerhoas L, Lettere M, Jouanneau Y, Marchal R (2008) Biodegradation of 2-ethylhexyl nitrate by Mycobacterium austroafricanum IFP 2173. Environ Microbiol 74:6187–6193
Ramirez KS, Craine JM, Fierer N (2012) Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Glob Chang Biol 18(6):1918–1927
Röling WF, Milner MG, Jones DM, Lee K, Daniel F, Swannell RJ, Head IM (2002) Robust hydrocarbon degradation and dynamics of bacterial communities during nutrient-enhanced oil spill bioremediation. Appl Environ Microbiol 68(11):5537–5548
Sambrook J, Russell DW (2001) Molecular cloning. A laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Shim H, Hwang B, Lee SS, Kong SH (2005) Kinetics of BTEX biodegradation by a coculture of Pseudomonas putida and Pseudomonas fluorescens under hypoxic conditions. Biodegradation 16(4):319–327
Smith RJ, Jeffries TC, Adetutu EM, Fairweather PG, Mitchell JG (2013) Determining the metabolic footprints of hydrocarbon degradation using multivariate analysis. PLoS One 8(11):e81910
Solano-Serena F, Marchal R, Casaregola S, Vasnier C, Lebeault JM, Vandecasteele J-P (2000) A Mycobacterium strain with extended capacities for degradation of gasoline hydrocarbons. Appl Environ Microbiol 66:2392–2399
Solano-Serena F, Marchal R, Heiss S, Vandecasteele J-P (2004) Degradation of isooctane by Mycobacterium austroafricanum IFP 2173: growth and catabolic pathway. J Appl Microbiol 629–639
Solano-Serena F, Marchal R, Vandecasteele J-P (2008) Biodegradation of aliphatic and alicyclic hydrocarbons. Petroleum microbiology: concepts. environmental implications. industrial applications. Editions Technip Paris France 1:170–240
Takeda H, Shimodaira J, Yukawa K, Hara N, Kasai D, Miyauchi K, Masai E, Fukuda M (2010) Dual two-component regulatory systems are involved in aromatic compound degradation in a polychlorinated-biphenyl degrader, Rhodococcus jostii RHA1. J Bacteriol 192(18):4741–4751
Thompson IP, van der Gast CJ, Ciric L, Singer AC (2005) Bioaugmentation for bioremediation: the challenge of strain selection. Environ Microbiol 7:909–915
Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI (2009) A core gut microbiome in obese and lean twins. Nature 457:480–484
Van Beilen JB, Panke S, Lucchini S, Franchini AG, Röthlisberger M, Witholt B (2001) Analysis of Pseudomonas putida alkane-degradation gene clusters and flanking insertion sequences: evolution and regulation of the alk genes. Microbiology 147:1621–1630
Van Hamme JD, Singh A, Ward OP (2003) Recent advances in petroleun microbiology. Microbiol Mol Biol Rev 67(4):503–549
Vandecasteele J-P, Monot F (2008) Biodegradation of monoaromatic and chloroaromatic hydrocarbons. In: Vandecasteele J-P (ed) Petroleum microbiology: concepts, environmental implications, industrial applications. Editions Technip, Paris, pp 240–339
Vinas M, Sabaté S, Espuny MJ, Solanas AM (2005) Bacterial community dynamics and polycyclic aromatic hydrocarbon degradation during bioremediation of heavily creosote-contaminated soil. Appl Environ Microbiol 71(11):7008–7018
Wang B, Lai Q, Cui Z, Tan T, Shao Z (2008) A pyrene-degrading consortium from deep-sea sediment of the west pacific and its key member Cycloclasticus sp. P1. Environ Microbiol 10(8):1948–1963
Whyte LG, Bourbonnière L, Greer CW (1997) Biodegradation of petroleum hydrocarbons by psychrotrophic Pseudomonas strains possessing both alkane (alk) and naphthalene (nah) catabolic pathways. Appl Environ Microbiol 63:3719–3723
Wu JH, Wu FY, Chuang HP, Chen WY, Huang HJ, Chen SH, Liu WT (2013) Community and proteomic analysis of methanogenic consortia degrading terephthalate. Appl Environ Microbiol 79(1):105–112
Yeung CW, Woo M, Lee K, Greer CW (2011) Characterization of the bacterial communitystructure of Sydney Tar Ponds sediment. Can J Microbiol 57(6):493–503
Yu K, Zhang T (2012) Metagenomic and metatranscriptomic analysis of microbial community structure and gene expression of activated sludge. PLoS One 7(5):e38183
Acknowledgments
Marc D. Auffret was partly supported by a CIFRE (Convention Industrielle de Formation par la Recherche) fellowship provided by ANRT (Association Nationale de la Recherche Technique) and by IFP. We thank Isabelle Durand and Françoise Le Roux for hydrocarbon analyses.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 139 kb)
Rights and permissions
About this article
Cite this article
Auffret, M.D., Yergeau, E., Labbé, D. et al. Importance of Rhodococcus strains in a bacterial consortium degrading a mixture of hydrocarbons, gasoline, and diesel oil additives revealed by metatranscriptomic analysis. Appl Microbiol Biotechnol 99, 2419–2430 (2015). https://doi.org/10.1007/s00253-014-6159-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-6159-8