Abstract
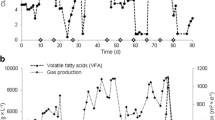
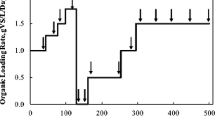
Droplet digital PCR (ddPCR) is a new DNA quantification platform without an external DNA calibrator. This study examined methanogen communities in four full-scale anaerobic digesters treating municipal sewage sludge, using ddPCR with taxon-specific primer/TaqMan probe sets (5 orders, 11 families, and 13 genera), many of which were developed in this study. Total methanogen abundance was positively correlated with hydraulic retention time (HRT) and temperature (p < 0.05), though the effect of HRT was stronger (r = 0.864 vs. 0.682, respectively). Moreover, total abundance was strongly correlated with biogas production rate (r = 0.896). HRT was positively correlated with seven methanogenic taxa, while temperature was positively or negatively correlated with 13 taxa (p < 0.05). For instance, the predominant genera Methanosaeta and Methanosarcina were negatively and positively associated, respectively, with temperature only (p < 0.05). Redundancy analysis and principal component analysis using the absolute-abundance dataset indicated that only temperature explained the variability in the methanogen communities at all classification levels. Therefore, HRT was the most important operational factor to influence net methanogen abundance and activity, while temperature governed the composition of the methanogen community. ddPCR enabled absolute quantification of methanogens without the external DNA standards and linked methanogen communities and operational factors, suggesting that it is a promising tool for analyzing the microbial ecology of anaerobic digestion.





Similar content being viewed by others
References
Blume F, Bergmann I, Nettmann E, Schelle H, Rehde G, Mundt K, Klocke M (2010) Methanogenic population dynamics during semi-continuous biogas fermentation and acidification by overloading. J Appl Microbiol 109:441–450
Brankatschk R, Bodenhausen N, Zeyer J, Bürgmann H (2012) Simple absolute quantification method correcting for quantitative PCR efficiency variations for microbial community samples. Appl Environ Microbiol 78:4481–4489
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM (2009) The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145
Demirel B, Scherer P (2008) The roles of acetotrophic and hydrogenotrophic methanogens during anaerobic conversion of biomass to methane: a review. Rev Environ Sci Biotechnol 7:173–190
Ferry JG, Smith PH, Wolfe RS (1974) Methanospirillum, a new genus of methanogenic bacteria, and characterization of Methanospirillum hungatii sp. nov. Int J Syst Bacteriol 24:465–469
Garcia J-L, Patel BKC, Ollivier B (2000) Taxonomic, phylogenetic, and ecological diversity of methanogenic archaea. Anaerobe 6:205–226
González JM, Masuchi Y, Robb FT, Ammerman JW, Maeder DL, Yanagibayashi M, Tamaoka J, Kato C (1998) Pyrococcus horikoshii sp. nov., a hyperthermophilic archaeon isolated from a hydrothermal vent at the Okinawa Trough. Extremophiles 2:123–130
Griffin ME, McMahon KD, Mackie RI, Raskin L (1998) Methanogenic population dynamics during start-up of anaerobic digesters treating municipal solid waste and biosolids. Biotechnol Bioeng 57:342–355
Hayden RT, Gu Z, Ingersoll J, Abdul-Ali D, Shi L, Pounds S, Caliendo AM (2013) Comparison of droplet digital PCR to real-time PCR for quantitative detection of Cytomegalovirus. J Clin Microbiol 51:540–546
Henrich TJ, Gallien S, Li JZ, Pereyra F, Kuritzkes DR (2012) Low-level detection and quantitation of cellular HIV-1 DNA and 2-LTR circles using droplet digital PCR. J Virol Methods 186:68–72
Higuchi R, Fockler C, Dollinger G, Watson R (1993) Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Nat Biotechnol 11:1026–1030
Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, Kitano TK, Hodel MR, Petersen JF, Wyatt PW, Steenblock ER, Shah PH, Bousse LJ, Troup CB, Mellen JC, Wittmann DK, Erndt NG, Cauley TH, Koehler RT, So AP, Dube S, Rose KA, Montesclaros L, Wang S, Stumbo DP, Hodges SP, Romine S, Milanovich FP, White HE, Regan JF, Karlin-Neumann GA, Hindson CM, Saxonov S, Colston BW (2011) High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 83:8604–8610
Ho DP, Jensen PD, Batstone DJ (2013) Methanosarcinaceae and acetate-oxidizing pathways dominate in high-rate thermophilic anaerobic digestion of waste-activated sludge. Appl Environ Microbiol 79:6491–6500
Ishii K, Fukui M (2001) Optimization of annealing temperature to reduce bias caused by a primer mismatch in multitemplate PCR. Appl Environ Microbiol 67:3753–3755
Kalendar R, Lee D, Schulman AH (2011) Java web tools for PCR, in silico PCR, and oligonucleotide assembly and analysis. Genomics 98:137–144
Karakashev D, Batstone DJ, Angelidaki I (2005) Influence of environmental conditions on methanogenic compositions in anaerobic biogas reactors. Appl Environ Microbiol 71:331–338
Kelley K, Cosman A, Belgrader P, Chapman B, Sullivan DC (2013) Detection of methicillin-resistant Staphylococcus aureus by a duplex droplet digital PCR assay. J Clin Microbiol 51:2033–2039
Kim TG, Jeong S-Y, Cho K-S (2014) Comparison of droplet digital PCR and quantitative real-time PCR for examining population dynamics of bacteria in soil. Appl Microbiol Biotechnol 98:6105–6113
Kim TG, Knudsen GR (2008) Quantitative real-time PCR effectively detects and quantifies colonization of sclerotia of Sclerotinia sclerotiorum by Trichoderma spp. Appl Soil Ecol 40:100–108
Kim TG, Yi T, Cho K-S (2013) Use of artificial DNA with multiple probe sites as reference DNA templates for quantitative real-time PCR to examine methanogen communities. J Environ Sci Health Part A 48:417–421
Klappenbach JA, Saxman PR, Cole JR, Schmidt TM (2001) rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res 29:181–184
Krakat N, Schmidt S, Scherer P (2010a) Mesophilic fermentation of renewable biomass: does hydraulic retention time regulate methanogen diversity? Appl Environ Microbiol 76:6322–6326
Krakat N, Westphal A, Schmidt S, Scherer P (2010b) Anaerobic digestion of renewable biomass: thermophilic temperature governs methanogen population dynamics. Appl Environ Microbiol 76:1842–1850
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and clustal X version 2.0. Bioinformatics 23:2947–2948
Laurie MT, Bertout JA, Taylor SD, Burton JN, Shendure JA, Bielas JH (2013) Simultaneous digital quantification and fluorescence-based size characterization of massively parallel sequencing libraries. Biotechniques 55:61–67
Lavoie I, Hamilton P, Poulin M (2011) Phytoplankton community metrics based on absolute and relative abundance and biomass: implications for multivariate analyses. J Appl Phycol 23:735–743
Leclerc M, Delgènes J-P, Godon J-J (2004) Diversity of the archaeal community in 44 anaerobic digesters as determined by single strand conformation polymorphism analysis and 16S rDNA sequencing. Environ Microbiol 6:809–819
Levén L, Eriksson ARB, Schnürer A (2007) Effect of process temperature on bacterial and archaeal communities in two methanogenic bioreactors treating organic household waste. FEMS Microbiol Ecol 59:683–693
Miroshnichenko ML, Gongadze GM, Rainey FA, Kostyukova AS, Lysenko AM, Chernyh NA, Bonch-Osmolovskaya EA (1998) Thermococcus gorgonarius sp. nov. and Thermococcus pacificus sp. nov.: heterotrophic extremely thermophilic archaea from New Zealand submarine hot vents. Int J Syst Bacteriol 48:23–29
Montero B, Garcia-Morales JL, Sales D, Solera R (2008) Evolution of microorganisms in thermophilic-dry anaerobic digestion. Bioresource Technol 99:3233–3243
Morisset D, Štebih D, Milavec M, Gruden K, Žel J (2013) Quantitative analysis of food and feed samples with droplet digital PCR. PLoS One 8:e62583
Narihiro T, Terada T, Ohashi A, Wu J-H, Liu W-T, Araki N, Kamagata Y, Nakamura K, Sekiguchi Y (2009) Quantitative detection of culturable methanogenic archaea abundance in anaerobic treatment systems using the sequence-specific rRNA cleavage method. ISME J 3:522–535
Pinheiro LB, Coleman VA, Hindson CM, Herrmann J, Hindson BJ, Bhat S, Emslie KR (2012) Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem 84:1003–1011
Reavie ED, Jicha TM, Angradi TR, Bolgrien DW, Hill BH (2010) Algal assemblages for large river monitoring: comparison among biovolume, absolute and relative abundance metrics. Ecol Indic 10:167–177
Rivière D, Desvignes V, Pelletier E, Chaussonnerie S, Guermazi S, Weissenbach J, Li T, Camacho P, Sghir A (2009) Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. ISME J 3:700–714
Sakai S, Imachi H, Hanada S, Ohashi A, Harada H, Kamagata Y (2008) Methanocella paludicola gen. nov., sp. nov., a methane-producing archaeon, the first isolate of the lineage ‘Rice Cluster I’ and proposal of the new archaeal order Methanocellales ord. nov. Int J Syst Evol Microbiol 58:929–936
Sawayama S, Tsukahara K, Yagishita T (2006) Phylogenetic description of immobilized methanogenic community using real-time PCR in a fixed-bed anaerobic digester. Bioresource Technol 97:69–76
Smith CJ, Osborn AM (2009) Advantages and limitations of quantitative PCR (Q-PCR)-based approaches in microbial ecology. FEMS Microbiol Ecol 67:6–20
Steinberg LM, Regan JM (2009) mcrA-targeted real-time quantitative PCR method to examine methanogen communities. Appl Environ Microbiol 75:4435–4442
Strain MC, Lada SM, Luong T, Rought SE, Gianella S, Terry VH, Spina CA, Woelk CH, Richman DD (2013) Highly precise measurement of HIV DNA by droplet digital PCR. PLoS One 8:e55943
Stults JR, Snoeyenbos-West O, Methe B, Lovley DR, Chandler DP (2001) Application of the 5′ fluorogenic exonuclease assay (TaqMan) for quantitative ribosomal DNA and rRNA analysis in sediments. Appl Environ Microbiol 67:2781–2789
Sundberg C, Al-Soud WA, Larsson M, Alm E, Yekta SS, Svensson BH, Sørensen SJ, Karlsson A (2013) 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol Ecol 85:612–626
Takai K, Horikoshi K (2000) Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl Environ Microbiol 66:5066–5072
ter Braak CJF, Šmilauer P (2002) CANOCO reference manual and CanoDraw for Windows user’s guide: software for canonical community ordination (version 4.5). Microcomputer Power, Ithaca, NY, pp 1–500
Tian J, Wang Y, Dong X (2010) Methanoculleus hydrogenitrophicus sp. nov., a methanogenic archaeon isolated from wetland soil. Int J Syst Evol Microbiol 60:2165–2169
Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden T (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform 13:134
Yu Y, Lee C, Kim J, Hwang S (2005) Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol Bioeng 89:670–679
Zielińska M, Cydzik-Kwiatkowska A, Zieliński M, Dębowski M (2013) Impact of temperature, microwave radiation and organic loading rate on methanogenic community and biogas production during fermentation of dairy wastewater. Bioresource Technol 129:308–314
Acknowledgments
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (NRF-2012R1A2A2A03046724).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kim, T.G., Jeong, SY. & Cho, KS. Development of droplet digital PCR assays for methanogenic taxa and examination of methanogen communities in full-scale anaerobic digesters. Appl Microbiol Biotechnol 99, 445–458 (2015). https://doi.org/10.1007/s00253-014-6007-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-6007-x