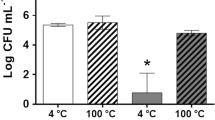
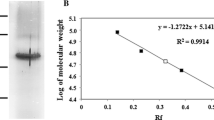
Abstract
Saccharomyces cerevisiae plays a primordial role in alcoholic fermentation and has a vast worldwide application in the production of fuel-ethanol, food and beverages. The dominance of S. cerevisiae over other microbial species during alcoholic fermentations has been traditionally ascribed to its higher ethanol tolerance. However, recent studies suggested that other phenomena, such as microbial interactions mediated by killer-like toxins, might play an important role. Here we show that S. cerevisiae secretes antimicrobial peptides (AMPs) during alcoholic fermentation that are active against a wide variety of wine-related yeasts (e.g. Dekkera bruxellensis) and bacteria (e.g. Oenococcus oeni). Mass spectrometry analyses revealed that these AMPs correspond to fragments of the S. cerevisiae glyceraldehyde 3-phosphate dehydrogenase (GAPDH) protein. The involvement of GAPDH-derived peptides in wine microbial interactions was further sustained by results obtained in mixed cultures performed with S. cerevisiae single mutants deleted in each of the GAPDH codifying genes (TDH1-3) and also with a S. cerevisiae mutant deleted in the YCA1 gene, which codifies the apoptosis-involved enzyme metacaspase. These findings are discussed in the context of wine microbial interactions, biopreservation potential and the role of GAPDH in the defence system of S. cerevisiae.






Similar content being viewed by others
References
Albergaria H, Francisco D, Gori K, Arneborg N, Gírio F (2010) Saccharomyces cerevisiae CCMI 885 secretes peptides that inhibit the growth of some non-Saccharomyces wine-related strains. Appl Microbiol Biotechnol 86:965–972. doi:10.1007/s00253-009-2409-6
Arneborg N, Siegumfeldt H, Andersen GH, Nissen P, Daria VR, Rodrigo PJ, Glückstad J (2005) Interactive optical trapping shows that confinement is a determinant of growth in a mixed yeast culture. FEMS Microbiol Lett 245:155–159. doi:10.1016/j.femsle.2005.03.008
Barata A, Caldeira J, Botelheiro R, Pagliara D, Malfeito-Ferreira M, Loureiro V (2008) Survival patterns of Dekkera bruxellensis in wines and inhibitory effect of sulphur dioxide. Int J Food Microbiol 121:201–207. doi:10.1016/j.ijfoodmicro.2007.11.020
Basílio ACM, Araújo PCM, Morais JOF, Silva-Filho EA, Morais MA, Simões DA (2008) Detection and identification of wild yeast contaminants of the industrial fuel ethanol fermentation process. Curr Microbiol 56:322–226. doi:10.1007/s00284-007-9085-5
Bauer FF, Pretorius IS (2000) Yeast stress response and fermentation efficiency: how to survive the making of wine—a review. S Afr J Enol Vitic 21:27–51
Bisson LF (1999) Stuck and sluggish fermentations. Am J Enol Vitic 50:107–119
Comitini F, Ciani M (2011) Kluyveromyces wickerhamii killer toxin: purification and activity towards Brettanomyces/Dekkera yeasts in grape must. FEMS Microbiol Lett 316:77–82. doi:10.1111/j.1574-6968.2004.tb09761.x
Comitini F, De JI, Pepe L, Mannazzu I, Ciani M (2004) Pichia anomala and Kluyveromyces wickerhamii killer toxins as new tools against Dekkera/Brettanomyces spoilage yeasts. FEMS Microbiol Lett 238:235–240. doi:10.1016/j.femsle.2004.07.040
Comitini F, Ferretti R, Clementi F, Mannuzzu I, Ciani M (2005) Interactions between Saccharomyces cerevisiae and malolactic bacteria: preliminary characterization of a yeast proteinaceous compound(s) active against Oenococcus oeni. J Appl Microbiol 99:105–111. doi:10.1111/j.1365-2672.2005.02579.x
Delgado ML, O’Connor JE, Azorin I, Renau-Piqueras J, Gil ML, Gozalbo D (2001) The glyceraldehyde-3-phosphate dehydrogenase polypeptides encoded by the Saccharomyces cerevisiae TDH1, TDH2 and TDH3 genes are also cell wall proteins. Microbiol 147:411–417
Delgado ML, Gil ML, Gozalbo D (2003) Starvation and temperature upshift cause an increase in the enzymaticaly active cell wall-associated glyceraldehyde-3-phosphate dehydrogenase protein in yeast. FEMS Yeast Res 4:297–303. doi:10.1016/S1567-1356(03)00159-4
Fleet GH, Heard GM (1993) Yeast growth during fermentation. In: Fleet GH (ed) Wine microbiology and biotechnology. Harwood Academic Publishers, Basel, Switzerland, pp 27–54
Hansen EH, Nissen P, Sommer P, Nielsen JC, Arneborg N (2001) The effect of oxygen on the survival of non-Saccharomyces yeasts during mixed culture fermentations of grape juice with Saccharomyces cerevisiae. J Appl Microbiol 91:541–547. doi:10.1046/j.1365-2672.2001.01426.x
Liberal AT, Basílio ACM, Resende AM, Brasileiro BTV, Silva-Filho EA, Morais JOF, Simões DA, Morais MA (2007) Identification of Dekkera bruxellensis as a major contaminant yeast in continuous fuel ethanol fermentation. J Appl Microbiol 102:538–547. doi:10.1111/j.1365-2672.2006.03082.x
Loureiro V, Malfeito-Ferreira M (2003) Spoilage yeasts in the wine industry (review). Int J Food Microbiol 86:23–50. doi:10.1016/S0168-1605(03)00246-0
McAlister L, Holland MJ (1985a) Isolation and characterization of yeast strains carrying mutations in the glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem 260:15013–15018
McAlister L, Holland MJ (1985b) Differential expression of the three yeast glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem 260:15019–15027
Nehme N, Mathieu F, Taillandier P (2010) Impact of the co-culture of Saccharomyces cerevisiae–Oenococcus oeni on malolactic fermentation and partial characterization of a yeast-derived inhibitory peptidic fraction. Food Microbiol 27:150–157. doi:10.1016/j.fm.2009.09.008
Nakajima H, Amano W, Kubo T, Fukuhara A, Ihara H, Azuma YT, Tajima H, Inui T, Sawa A, Takeuchi T (2009) Glyceraldehyde-3-phosphate dehydrogenase aggregate formation participates in oxidative stress-induced cell death. J Cell Biochem 284:34331–34341. doi:10.1074/jbc.M109.027698
Nissen P, Arneborg N (2003) Characterization of early deaths of non-Saccharomyces yeasts in mixed cultures with Saccharomyces cerevisiae. Arch Microbiol 180:257–263. doi:10.1007/s00203-003-0585-9
Nissen P, Nielsen D, Arneborg N (2003) Viable Saccharomyces cerevisiae cells at high concentrations cause early growth arrest of non-Saccharomyces yeasts in mixed cultures by a cell-cell contact-mediated mechanism. Yeast 20:331–341. doi:10.1002/yea.965
Osborne JP, Edwards CG (2007) Inhibition of malolactic fermentation by a peptide produced by Saccharomyces cerevisiae during alcoholic fermentation. Int J Food Microbiol 118:27–34. doi:10.1016/j.ijfoodmicro.2007.05.007
Pérez F, Ramírez M, Regodón JA (2001) Influence of killer strains of Saccharomyces cerevisiae on wine fermentation. Antonie Van Leeuwenhoek 79:393–399. doi:10.1023/A:1012034608908
Pérez-Nevado F, Albergaria H, Hogg T, Gírio F (2006) Cellular death of two non-Saccharomyces wine-related yeasts during mixed fermentations with Saccharomyces cerevisiae. Int J Food Microbiol 108:336–345. doi:10.1016/j.ijfoodmicro.2005.12.012
Pretorius IS (2000) Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast 16:675–729. doi:10.1002/1097-0061(20000615)16:8<675::AID-YEA585>3.0.CO;2-B
Seo J-K, Lee MJ, Go H-J, Tae Hyun Park TH, Park NG (2012) Purification and characterization of YFGAP, a GAPDH-related novel antimicrobial peptide, from the skin of yellowfin tuna, Thunnus albacores. Fish & Shellfish Immunol 33:743–752. doi:10.1016/j.fsi.2012.06.023
Silva A, Almeida B, Sampaio-Marques B, Reis MIR, Ohlmeier S, Rodrigues F, Do Vale A, Ludovico P (2011) Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a specific substrate of yeast metacaspase. Biochem Biophys Acta 1813:2044–2049. doi:10.1016/j.bbamcr.2011.09.010
Sirover MA (2005) New nuclear functions of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in mammalian cells. J Cell Biochem 95:45–52. doi:10.1002/jcb.20399
Wagener J, Schneider JJ, Baxmann S, Kalbacher H, Borelli C, Nuding S, Küchler R, Wehkamp J, Kaeser MD, Mailänder-Sanchez D, Braunsdorf C, Hube B, Schild L, Forssmann W-G, Korting H-C, Liepke C, Schaller M (2013) A peptide derived from the highly conserved protein GAPDH is involved in tissue protection by different antifungal strategies and epithelial immunomodulation. J Investigat Dermatol 133:144–153. doi:10.1038/jid.2012.254
Acknowledgments
The present work was financed by FEDER funds through POFC-COMPETE and by national funds through Fundação para a Ciência e a Tecnologia (FCT) in the scope of project FCOMP-01-0124-FEDER-014055. M.G.A. and J.C. acknowledge the funding support from FCT (PEst-C/EQB/LA0006/2011). Patrícia Branco is the recipient of a PhD fellowship (SFRH/BD/89673/2012) funded by FCT, Portugal. We want also to thanks to Professors Isabel Sá-Correia (IST/UTL, Lisbon, Portugal), Luísa Marinho (FCUL, Lisbon, Portugal) and Paula Ludovico (ICVS/UM, Braga, Portugal) for kindly providing some mutant strains.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 84 kb)
Rights and permissions
About this article
Cite this article
Branco, P., Francisco, D., Chambon, C. et al. Identification of novel GAPDH-derived antimicrobial peptides secreted by Saccharomyces cerevisiae and involved in wine microbial interactions. Appl Microbiol Biotechnol 98, 843–853 (2014). https://doi.org/10.1007/s00253-013-5411-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-013-5411-y