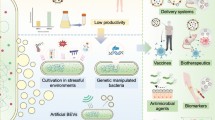
Abstract
Gram-positive bacteria are widely used to produce recombinant proteins, amino acids, organic acids, higher alcohols, and polymers. Many proteins have been expressed in Gram-positive hosts such as Corynebacterium, Brevibacterium, and Streptomyces. The favorable and advantageous characteristics (e.g., high secretion capacity and efficient production of metabolic products) of these species have increased the biotechnological applications of bacteria. However, owing to multiplicity from genes encoding the proteins and expression hosts, the expression of recombinant proteins is limited in Gram-positive bacteria. Because there is a very recent review about protein expression in Bacillus subtilis, here we summarize recent strategies for efficient expression of recombinant proteins in the other three typical Gram-positive bacteria (Corynebacterium, Brevibacterium, and Streptomyces) and discuss future prospects. We hope that this review will contribute to the development of recombinant protein expression in Corynebacterium, Brevibacterium, and Streptomyces.

Similar content being viewed by others
References
Adham SAI, Campelo AB, Ramos A, Gil JA (2001a) Construction of a xylanase-producing strain of Brevibacterium lactofermentum by stable integration of an engineered xysA gene from Streptomyces halstedii JM8. App Environ Microbiol 67:5425–5430
Adham SAI, Honrubia P, Diaz M, Fernandez-Abalos JM, Santamaria RI, Gil JA (2001b) Expression of the genes coding for the xylanase Xys1 and the cellulase Cel1 from the straw-decomposing Streptomyces halstedii JM8 cloned into the amino-acid producer Brevibacterium lactofermentum ATCC13869. Arch Microbiol 177:91–97
Anne J, Maldonado B, Van Impe J, Van Mellaert L, Bernaerts K (2012) Recombinant protein production and streptomycetes. J Biotechnol 158:159–167
Anne J, Mellaert LV (1993) Streptomyces lividans as host for heterologous protein production. FEMS Microbiol Lett 114:121–128
Azza S, Bigey F, Arnaud A, Galzy P (1994) Cloning of the wide spectrum amidase gene from Brevibacterium sp. R312 by genetic complementation. Overexpression in Brevibacterium sp. and Escherichia coli. FEMS Microbiol Lett 122:129–136
Becker J, Klopprogge C, Herold A, Zelder O, Bolten CJ, Wittmann C (2007) Metabolic flux engineering of l-lysine production in Corynebacterium glutamicum—over expression and modification of G6P dehydrogenase. J Biotechnol 132:99–109
Becker J, Zelder O, Haefner S, Schroeder H, Wittmann C (2011) From zero to hero-design-based systems metabolic engineering of Corynebacterium glutamicum for l-lysine production. Metab Eng 13:159–168
Berks BC, Palmer T, Sargent F (2003) The Tat protein translocation pathway and its role in microbial physiology. Adv Microb Physiol 47:187–254
Bibb MJ, Janssen GR, Ward JM (1985) Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene 38:215–226
Binnie C, Cossar JD, Stewart DI (1997) Heterologous biopharmaceutical protein expression in Streptomyces. Trends Biotechnol 15:315–320
Blombach B, Riester T, Wieschalka S, Ziert C, Youn J-W, Wendisch VF, Eikmanns BJ (2011) Corynebacterium glutamicum tailored for efficient isobutanol production. Appl Environ Microbiol 77:3300–3310
Bussmann M, Emer D, Hasenbein S, Degraf S, Eikmanns BJ, Bott M (2009) Transcriptional control of the succinate dehydrogenase operon sdhCAB of Corynebacterium glutamicum by the cAMP-dependent regulator GlxR and the LuxR-type regulator RamA. J Biotechnol 143:173–182
Butenas S (2013) Comparison of natural and recombinant tissue factor proteins: new insights. Biol Chem 394:819–829
Chater KF, Biro S, Lee KJ, Palmer T, Schrempf H (2010) The complex extracellular biology of Streptomyces. FEMS Microbiol Rev 34:171–198
Chauhan AK, Survase SA, Kishenkumar J, Annapure US (2009) Medium optimization by orthogonal array and response surface methodology for cholesterol oxidase production by Streptomyces lavendulae NCIM 2499. J Gen Appl Microbiol 55:171–180
Chen W, Qin Z (2011) Development of a gene cloning system in a fast-growing and moderately thermophilic Streptomyces species and heterologous expression of Streptomyces antibiotic biosynthetic gene clusters. BMC Microbiol 11:243
Chen Y, Smanski MJ, Shen B (2010) Improvement of secondary metabolite production in Streptomyces by manipulating pathway regulation. Appl Microbiol Biotechnol 86:19–25
Chion CK, Duran R, Arnaud A, Galzy P (1991) Cloning vectors and antibiotic-resistance markers for Brevibacterium sp. R312. Gene 105:119–124
Chubiz LM, Rao CV (2008) Computational design of orthogonal ribosomes. Nucleic Acids Res 36:4038–4046
Cramer A, Gerstmeir R, Schaffer S, Bott M, Eikmanns BJ (2006) Identification of RamA, a novel LuxR-type transcriptional regulator of genes involved in acetate metabolism of Corynebacterium glutamicum. J Bacteriol 188:2554–2567
Date M, Itaya H, Matsui H, Kikuchi Y (2006) Secretion of human epidermal growth factor by Corynebacterium glutamicum. Lett Appl Microbiol 42:66–70
Diaz M, Adham SAI, Ramon D, Gil JA, Santamaria RI (2004) Streptomyces lividans and Brevibacterium lactofermentum as heterologous hosts for the production of X-22 xylanase from Aspergillus nidulans. Appl Microbiol Biotechnol 65:401–406
Diaz M, Ferreras E, Moreno R, Yepes A, Berenguer J, Santamaria R (2008) High-level overproduction of Thermus enzymes in Streptomyces lividans. Appl Microbiol Biotechnol 79:1001–1008
Ehira S, Ogino H, Teramoto H, Inui M, Yukawa H (2009) Regulation of quinone oxidoreductase by the redox-sensing transcriptional regulator QorR in Corynebacterium glutamicum. J Biol Chem 284:16736–16742
Fang BS, Li W, Ng IS, Yu JC, Zhang GY (2011) Codon optimization of 1,3-propanediol oxidoreductase expression in Escherichia coli and enzymatic properties. Electron J Biotechn 14(4). doi:10.2225/vol14-issue4-fulltext-9
Fernandez M, Sanchez J (2001) Viability staining and terminal deoxyribonucleotide transferase-mediated dUTP nick end labelling of the mycelium in submerged cultures of Streptomyces antibioticus ETH7451. J Microbiol Methods 47:293–298
Forquin M-P, Hebert A, Roux A, Aubert J, Proux C, Heilier J-F, Landaud S, Junot C, Bonnarme P, Martin-Verstraete I (2011) Global regulation of the response to sulfur availability in the cheese-related bacterium Brevibacterium aurantiacum. Appl Environ Microbiol 77:1449–1459
Gerstmeir R, Cramer A, Dangel P, Schaffer S, Eikmanns BJ (2004) RamB, a novel transcriptional regulator of genes involved in acetate metabolism of Corynebacterium glutamicum. J Bacteriol 186:2798–2809
Han R, Li J, H-d S, Chen RR, Du G, Liu L, Chen J (2013) Carbohydrate-binding module fusion with cyclodextrin glycosyltransferase enables the efficient synthesis of 2-Od-glucopyranosyl-l-ascorbic acid with soluble starch as the glycosyl donor. Appl Environ Microbiol 79:3234–3240
Herai S, Hashimoto Y, Higashibata H, Maseda H, Ikeda H, Omura S, Kobayashi M (2004) Hyper-inducible expression system for Streptomycetes. Proc Natl Acad Sci USA 101:14031–14035
Hou X, Ge X, Wu D, Qian H, Zhang W (2012) Improvement of l-valine production at high temperature in Brevibacterium flavum by overexpressing ilvEBN(r)C genes. J Ind Microbiol Biotechnol 39:63–72
Hou Y, Wang H, Wang QL, Zhang FF, Huang YH, Ji YL (2008) Protein expression and purification of human Zbtb7A in Pichia pastoris via gene codon optimization and synthesis. Protein Expr Purif 60:97–102
Hyeon JE, Kang DH, Kim YI, You SK, Han SO (2012) GntR-type transcriptional regulator PckR negatively regulates the expression of phosphoenolpyruvate carboxykinase in Corynebacterium glutamicum. J Bacteriol 194:2181–2188
Ikeda M, Mizuno Y, S-i A, Hayashi M, Mitsuhashi S, Takeno S (2011) Identification and application of a different glucose uptake system that functions as an alternative to the phosphotransferase system in Corynebacterium glutamicum. Appl Microbiol Biotechnol 90:1443–1451
Isaacs FJ, Dwyer DJ, Ding C, Pervouchine DD, Cantor CR, Collins JJ (2004) Engineered riboregulators enable post-transcriptional control of gene expression. Nat Biotechnol 22:841–847
Itaya H, Kikuchi Y (2008) Secretion of Streptomyces mobaraensis pro-transglutaminase by coryneform bacteria. Appl Microbiol Biotechnol 78:621–625
Jones AC, Ottilie S, Eustaquio AS, Edwards DJ, Gerwick L, Moore BS, Gerwick WH (2012) Evaluation of Streptomyces coelicolor A3(2) as a heterologous expression host for the cyanobacterial protein kinase C activator lyngbyatoxin A. FEBS J 279:1243–1251
Jong-Uk P, Jo J-H, Kim Y-J, Chung S-S, Lee J-H, Lee HH (2008) Construction of heat-inducible expression vector of Corynebacterium glutamicum and C. ammoniagenes: fusion of lambda operator with promoters isolated from C. ammoniagenes. J Microbiol Biotechnol 18:639–647
Kabus A, Georgi T, Wendisch VF, Bott M (2007) Expression of the Escherichia coli pntAB genes encoding a membrane-bound transhydrogenase in Corynebacterium glutamicum improves l-lysine formation. Appl Microbiol Biotechnol 75:47–53
Kawaguchi H, Sasaki M, Vertes AA, Inui M, Yukawa H (2009) Identification and functional analysis of the gene cluster for l-arabinose utilization in Corynebacterium glutamicum. Appl Environ Microbiol 75:3419–3429
Kikuchi Y, Date M, Itaya H, Matsui K, Wu L-F (2006) Functional analysis of the twin-arginine translocation pathway in Corynebacterium glutamicum ATCC 13869. Appl Environ Microbiol 72:7183–7192
Kikuchi Y, Date M, Yokoyama K, Umezawa Y, Matsui H (2003) Secretion of active-form Streptoverticillium mobaraense transglutaminase by Corynebacterium glutamicum: processing of the pro-transglutaminase by a cosecreted subtilisin-like protease from Streptomyces albogriseolus. Appl Environ Microbiol 69:358–366
Komatsu M, Uchiyama T, Omura S, Cane DE, Ikeda H (2010) Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc Natl Acad Sci USA 107:2646–2651
Krug A, Wendisch VF, Bott M (2005) Identification of AcnR, a TetR-type repressor of the aconitase gene acn in Corynebacterium glutamicum. J Biol Chem 280:585–595
Lammertyn E, Desmyter S, Schacht S, Van Mellaert L, Anne J (1998) Influence of charge variation in the Streptomyces venezuelae alpha-amylase signal peptide on heterologous protein production by Streptomyces lividans. Appl Microbiol Biotechnol 49:424–430
Lara M, Servin-Gonzalez L, Singh M, Moreno C, Cohen I, Nimtz M, Espitia C (2004) Expression, secretion, and glycosylation of the 45-and 47-kDa glycoprotein of Mycobacterium tuberculosis in Streptomyces lividans. Appl Environ Microbiol 70:679–685
Lausberg F, Chattopadhyay AR, Heyer A, Eggeling L, Freudl R (2012) A tetracycline inducible expression vector for Corynebacterium glutamicum allowing tightly regulable gene expression. Plasmid 68:142–147
Lee SK, Keasling JD (2006) A Salmonella-based, propionate-inducible, expression system for Salmonella enterica. Gene 377:6–11
Leskiw BK, Lawlor EJ, Fernandez-Abalos JM, Chater KF (1991) TTA codons in some genes prevent their expression in a class of developmental, antibiotic-negative, Streptomyces mutants. Proc Natl Acad Sci USA 88:2461–2465
Lin SJ, Hsieh YF, Lai LA, Chao ML, Chu WS (2008) Characterization and large-scale production of recombinant Streptoverticillium platensis transglutaminase. J Ind Microbiol Biotechnol 35:981–990
Lin SJ, Hsieh YF, Wang PM, Chu WS (2007) Efficient purification of transglutaminase from recombinant Streptomyces platensis at various scales. Biotechnol Lett 29:111–115
Lin YS, Chao ML, Liu CH, Chu WS (2004) Cloning and expression of the transglutaminase gene from Streptoverticillium ladakanum in Streptomyces lividans. Process Biochem 39:591–598
Lin YS, Chao ML, Liu CH, Tseng M, Chu WS (2006) Cloning of the gene coding for transglutaminase from Streptomyces platensis and its expression in Streptomyces lividans. Process Biochem 41:519–524
Litsanov B, Brocker M, Bott M (2012) Toward homosuccinate fermentation: metabolic engineering of Corynebacterium glutamicum for anaerobic production of succinate from glucose and formate. Appl Environ Microbiol 78:3325–3337
Liu L, Liu YF, H-d S, Chen RR, Wang NS, Li J, Du G, Chen J (2013) Developing Bacillus spp. as a cell factory for production of microbial enzymes and industrially important biochemicals in the context of systems and synthetic biology. Appl Microbiol Biotechnol 97:6113–6127
Low KO, Mahadi NM, Illias RM (2013) Optimization of signal peptide for recombinant protein secretion in bacterial hosts. Appl Microbiol Biotechnol 97:3811–3826
Lussier FX, Denis F, Shareck F (2010) Adaptation of the highly productive T7 expression system to Streptomyces lividans. Appl Environ Microbiol 76:967–970
Malumbers M (1993) Cloning and characterization of genes involved in threonine and lysine biosynthesis in Brevibacterium lactofermentum. Ph.D. thesis, University of León, Spain
Manteca A, Alvarez R, Salazar N (2008) Mycelium differentiation and antibiotic production in submerged cultures of Streptomyces coelicolor. Appl Environ Microbiol 74:3877–3886
Manteca A, Claessen D, Lopez-Iglesias C, Sanchez J (2007) Aerial hyphae in surface cultures of Streptomyces lividans and Streptomyces coelicolor originate from viable segments surviving an early programmed cell death event. FEMS Microbiol Lett 274:118–125
Manteca A, Sanchez J (2009) Streptomyces development in colonies and soils. Appl Environ Microbiol 75:2920–2924
McVey JH, Ward NJ, Buckley SMK, Waddington SN, VandenDriessche T, Chuah MKL, Nathwani AC, McIntosh J, Tuddenham EGD, Kinnon C, Thrasher AJ (2011) Codon optimization of human factor VIII cDNAs leads to high-level expression. Blood 117:798–807
Medema MH, Breitling R, Takano E (2011) Synthetic biology in Streptomyces bacteria. Method Enzymol 497:485–502
Medema MH, Trefzer A, Kovalchuk A, van den Berg M, Muller U, Heijne W, Wu LA, Alam MT, Ronning CM, Nierman WC, Bovenberg RAL, Breitling R, Takano E (2010) The sequence of a 1.8-Mb bacterial linear plasmid reveals a rich evolutionary reservoir of secondary metabolic pathways. Genome Biol Evol 2:212–224
Meissner D, Vollstedt A, van Dijl JM, Freudl R (2007) Comparative analysis of twin-arginine (Tat)-dependent protein secretion of a heterologous model protein (GFP) in three different Gram-positive bacteria. Appl Microbiol Biotechnol 76:633–642
Morosoli R, Dupont C (1999) Secretion of xylanase A2 in Streptomyces lividans: dependence on signal peptides length, number and composition. FEMS Microbiol Lett 179:437–445
Nakashima N, Mitani Y, Tamura T (2005) Actinomycetes as host cells for production of recombinant proteins. Microb Cell Fact 4:7
Nazari B, Saito A, Kobayashi M, Miyashita K, Wang Y, Fujii T (2011) High expression levels of chitinase genes in Streptomyces coelicolor A3(2) grown in soil. FEMS Microbiol Ecol 77:623–635
Nesvera J, Holatko J, Patek M (2012) Analysis of Corynebacterium glutamicum promoters and their applications. Sub-Cellular Biochem 64:203–221
Nguyen KT, Tenor J, Stettler H, Nguyen LT, Nguyen LD, Thompson CJ (2003) Colonial differentiation in Streptomyces coelicolor depends on translation of a specific codon within the adpA gene. J Bacteriol 185:7291–7296
Niimi S, Suzuki N, Inui M, Yukawa H (2011) Metabolic engineering of 1,2-propanediol pathways in Corynebacterium glutamicum. Appl Microbiol Biotechnol 90:1721–1729
Niladevi KN, Sukumaran RK, Jacob N, Anisha GS, Prema P (2009) Optimization of laccase production from a novel strain-Streptomyces psammoticus using response surface methodology. Microbiol Res 164:105–113
Ninawe S, Kapoor M, Kuhad RC (2008) Purification and characterization of extracellular xylanase from Streptomyces cyaneus SN32. Bioresour Technol 99:1252–1258
Noda S, Ito Y, Shimizu N, Tanaka T, Ogino C, Kondo A (2010) Overproduction of various secretory-form proteins in Streptomyces lividans. Protein Expr Purif 73:198–202
Okibe N, Suzuki N, Inui M, Yukawa H (2010) Isolation, evaluation and use of two strong, carbon source-inducible promoters from Corynebacterium glutamicum. Lett Appl Microbiol 50:173–180
Paradis FW, Warren RA, Kilburn DG, Miller RC Jr (1987) The expression of Cellulomonas fimi cellulase genes in Brevibacterium lactofermentum. Gene 61:199–206
Parks L (2012) Biopharmaceutical companies unite in production of antibody-drug conjugates. Bioanalysis 4:2871–2871
Patek M, Nesvera J (2011) Sigma factors and promoters in Corynebacterium glutamicum. J Biotechnol 154:101–113
Peters-Wendisch P, Stolz M, Etterich H, Kennerknecht N, Sahm H, Eggeling L (2005) Metabolic engineering of Corynebacterium glutamicum for l-serine production. Appl Environ Microbiol 71:7139–7144
Pimienta E, Ayala JC, Rodriguez C, Ramos A, Van Mellaert L, Vallin C, Anne J (2007) Recombinant production of Streptococcus equisimilis streptokinase by Streptomyces lividans. Microb Cell Fact 6:20
Plassmeier J, Persicke M, Puehler A, Sterthoff C, Rueckert C, Kalinowski J (2012) Molecular characterization of PrpR, the transcriptional activator of propionate catabolism in Corynebacterium glutamicum. J Biotechnol 159:1–11
Plassmeier JK, Busche T, Molck S, Persicke M, Puhler A, Ruckert C, Kalinowski J (2013) A propionate-inducible expression system based on the Corynebacterium glutamicum prpD2 promoter and PrpR activator and its application for the redirection of amino acid biosynthesis pathways. J Biotechnol 163:225–232
Rueda B, Miguelez EM, Hardisson C, Manzanal MB (2001) Mycelial differentiation and spore formation by Streptomyces brasiliensis in submerged culture. Can J Microbiol 47:1042–1047
Ryu JK, Kim HS, Nam DH (2012) Current status and perspectives of biopharmaceutical drugs. Biotechnol Bioprocess Eng 17:900–911
Salis HM, Mirsky EA, Voigt CA (2009) Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol 27:946–950
Sanchez S, Demain A (2012) Special issue on the production of recombinant proteins. Biotechnol Adv 30:1100–1101
Schaerlaekens K, Lammertyn E, Geukens N, De Keersmaeker S, Anne J, Van Mellaert L (2004) Comparison of the Sec and Tat secretion pathways for heterologous protein production by Streptomyces lividans. J Biotechnol 112:279–288
Schmitt-John T, Engels JW (1992) Promoter constructions for efficient secretion expression in Streptomyces lividans. Appl Microbiol Biotechnol 36:493–498
Shi F, Huan X, Wang X, Ning J (2012) Overexpression of NAD kinases improves the l-isoleucine biosynthesis in Corynebacterium glutamicum ssp lactofermentum. Enzyme Microb Technol 51:73–80
Shin H-S, Kim Y-J, Yoo I-H, Lee H-S, Jin S, Ha U-H (2011) Autoinduction of a genetic locus encoding putative acyltransferase in Corynebacterium glutamicum. Biotechnol Lett 33:97–102
Song Y, Ki M, Yamada M, Gohda A, Brigham CJ, Sinskey AJ, Taguchi S (2012) Engineered Corynebacterium glutamicum as an endotoxin-free platform strain for lactate-based polyester production. Appl Microbiol Biotechnol 93:1917–1925
Srivastava P, Deb JK (2005) Gene expression systems in Corynebacteria. Protein Expre Purif 40:221–229
Strohl WR (1992) Compilation and analysis of DNA sequences associated with apparent Streptomycete promoters. Nucleic Acids Res 20:961–974
Takano E, White J, Thompson CJ, Bibb MJ (1995) Construction of thiostrepton-inducible, high-copy-number expression vectors for use in Streptomyces spp. Gene 166:133–137
Tanaka Y, Ehira S, Teramoto H, Inui M, Yukawa H (2012) Coordinated regulation of gnd, which encodes 6-phosphogluconate dehydrogenase, by the two transcriptional regulators GntR1 and RamA in Corynebacterium glutamicum. J Bacteriol 194:6527–6536
Tateno T, Fukuda H, Kondo A (2007a) Direct production of l-lysine from raw corn starch by Corynebacterium glutamicum secreting Streptococcus bovis alpha-amylase using cspB promoter and signal sequence. Appl Microbiol Biotechnol 77:533–541
Tateno T, Fukuda H, Kondo A (2007b) Production of l-lysine from starch by Corynebacterium glutamicum displaying alpha-amylase on its cell surface. Appl Microbiol Biotechnol 74:1213–1220
Tateno T, Okada Y, Tsuchidate T, Tanaka T, Fukuda H, Kondo A (2009) Direct production of cadaverine from soluble starch using Corynebacterium glutamicum coexpressing alpha-amylase and lysine decarboxylase. Appl Microbiol Biotechnol 82:115–121
Teramoto H, Watanabe K, Suzuki N, Inui M, Yukawa H (2011) High yield secretion of heterologous proteins in Corynebacterium glutamicum using its own Tat-type signal sequence. Appl Microbiol Biotechnol 91:677–687
Toyoda K, Teramoto H, Inui M, Yukawa H (2008) Expression of the gapA gene encoding glyceraldehyde-3-phosphate dehydrogenase of Corynebacterium glutamicum is regulated by the global regulator SugR. Appl Microbiol Biotechnol 81:291–301
Tsuchidate T, Tateno T, Tanaka T, Ogino C, Kondo A (2009) Engineering of endoglucanase expression system for Corynebacterium glutamicum. J Biosci Bioeng 108:S48–S49
Vrancken K, Anne J (2009) Secretory production of recombinant proteins by Streptomyces. Future Microbiol 4:181–188
Watanabe K, Tsuchida Y, Okibe N, Teramoto H, Suzuki N, Inui M, Yukawa H (2009) Scanning the Corynebacterium glutamicum R genome for high-efficiency secretion signal sequences. Microbiology-Sgm 155:741–750
Wennerhold J, Krug A, Bott M (2005) The AraC-type regulator RipA represses aconitase and other iron proteins from Corynebacterium under iron limitation and is itself repressed by DtxR. J Biol Chem 280:40500–40508
Wieschalka S, Blombach B, Eikmanns BJ (2012) Engineering Corynebacterium glutamicum for the production of pyruvate. Appl Microbiol Biotechnol 94:449–459
Wu YQ, Jiang PH, Fan CS, Wang JG, Shang L, Huang WD (2003) Co-expression of five genes in E. coli or l-phenylalanine in Brevibacterium flavum. World J Gastroenterology 9:342–346
Xu D, Tan Y, Huan X, Hu X, Wang X (2010) Construction of a novel shuttle vector for use in Brevibacterium flavum, an industrial amino acid producer. J Microbiol Methods 80:86–92
Xu D, Tan Y, Li Y, Wang X (2011) Construction of a novel promoter-probe vector and its application for screening strong promoter for Brevibacterium flavum metabolic engineering. World J Microbiol Biotechnol 27:961–968
Yague P, Manteca A, Simon A, Diaz-Garcia ME, Sanchez J (2010) A new method for monitoring programmed cell death and differentiation in submerged cultures of Streptomyces. Appl Environ Microbiol 76:3401–3404
Yamamoto S, Gunji W, Suzuki H, Toda H, Suda M, Jojima T, Inui M, Yukawa H (2012) Overexpression of genes encoding glycolytic enzymes in Corynebacterium glutamicum enhances glucose metabolism and alanine production under oxygen deprivation conditions. Appl Environ Microbiol 78:4447–4457
Yin L, Hu X, Xu D, Ning J, Chen J, Wang X (2012) Co-expression of feedback-resistant threonine dehydratase and acetohydroxy acid synthase increase l-isoleucine production in Corynebacterium glutamicum. Metab Eng 14:542–550
Zhang Y, Shang X, Lai S, Zhang G, Liang Y, Wen T (2012) Development and application of an arabinose-inducible expression system by facilitating inducer uptake in Corynebacterium glutamicum. Appl Environ Microbiol 78:5831–5838
Zhu YJ, Wang LF, Du Y, Wang SM, Yu TF, Hong B (2011) Heterologous expression of human interleukin-6 in Streptomyces lividans TK24 using novel secretory expression vectors. Biotechnol Lett 33:253–261
Zupancic TJ, Kittle JD, Baker BD, Miller CJ, Palmer DT, Asai Y, Inui M, Vertes A, Kobayashi M, Kurusu Y (1995) Isolation of promoters from Brevibacterium flavum strain MJ233C and comparison of their gene expression levels in B. flavum and Escherichia coli. FEMS Microbiol Lett 131:121–126
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Liu, L., Yang, H., Shin, Hd. et al. Recent advances in recombinant protein expression by Corynebacterium, Brevibacterium, and Streptomyces: from transcription and translation regulation to secretion pathway selection. Appl Microbiol Biotechnol 97, 9597–9608 (2013). https://doi.org/10.1007/s00253-013-5250-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-013-5250-x