Abstract
In this work, a three-level Salmonella detection system based on a combination of seven SYBR®Green qPCR was developed. This detection system discriminates Salmonella at the genus, species and subspecies levels using a single 96-well plate. The SYBR®Green qPCR assays target the invA, rpoD, iroB and safC genes, as well as the STM0296 locus, putatively coding for a cytoplasmic protein. This study includes the design of primer pairs, in silico and in situ selectivity, sensitivity, repeatability and reproducibility evaluations of the seven SYBR®Green qPCR assays. Each detection level displayed a selectivity of 100 %. This combinatory SYBR®Green qPCR system was also compared with three commercially available Salmonella qPCR detection kits. This comparison highlighted the importance of using a multi-gene detection system to be able to detect every target strain, even those with deletion or mutation of important genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The food safety is an important concern worldwide. Foodborne diseases cause not only an enormous economic burden due to sick leaves, treatments, hospitalisations and mortality (Scharff 2012) but also economical losses of farmers and industry. The recent outbreak of the O104:H4 serotype of Escherichia coli in Germany was a dramatic illustration of all the consequences of a severe outbreak (European Commission 2011). Currently, the official methods for the detection of pathogenic bacteria in food samples are norms from the International Organization for Standardization (ISO). These norms are culture-based methods. They are reliable but focus on individual pathogenic species, are time consuming and labour intensive (e.g. at least 5 days for Salmonella detection (ISO 6579 norm 2002)). Therefore, developing fast and simple foodborne pathogens detection tools, able to simultaneously detect several pathogens, remains a not only necessity but also a real challenge.
Real-time PCR (qPCR) assays have gained confidence as faster and reliable alternatives (Postollec et al. 2011), and several qPCR assays have already been developed (e.g. Beutin et al. 2009; Botteldoorn et al. 2003; Fukushima et al. 2009; Garrido et al. 2012b; Kim and Cho 2010; Löfström et al. 2010; Singh et al. 2012). However, their use all together into the same detection system is hampered by several drawbacks: they are using different technologies (e.g. TaqMan® and SYBR®Green), they are using different protocols and they were developed for different purposes and validated following different ways. Moreover, current detection assays are mostly based on the detection of a single virulence gene (e.g. invA for Salmonella or hlyA for Listeria detection). Yet, it has now been documented that Salmonella can lose an entire Pathogenicity island while remaining infectious (Hu et al. 2008). Single-gene target detection systems could, in this case, lead to a false negative signal.
In Europe, Salmonella is the first cause of notification of foodborne contamination due to microorganisms into the Rapid Alert System for Food and Feed (European Commission 2012) and the first detected causative agent in foodborne outbreaks reported, representing ca. 30 % of the total number of reported outbreaks in Europe in 2011 (European Food Safety Authority, European Centre for Disease Prevention and Control 2013). Salmonellosis is caused by the consumption of contaminated food (Gomez-Aldapa et al. 2012) (e.g. meat, raw eggs, cheese, fruit or vegetables) and, to a lesser extent, person-to-person transmission (Medus et al. 2006), waterborne transmission and environmental or animal exposure. The reservoirs are mainly poultry but also involve cattle, swine, sheep, birds, rats and reptiles (Bertrand et al. 2008; Pui et al. 2011). In developing countries, some Salmonella serovars can provoke typhoid and paratyphoid fevers. These serovars are not transmitted by food but mainly by drinking water spoiled by urine and faeces of infected individuals.
The Salmonella genus includes two species: Salmonella enterica and Salmonella bongori, both pathogenic for humans (Guibourdenche et al. 2010). The S. enterica species is itself divided into six subspecies: enterica, salamae, arizonae, diarizonae, houtenae and indica (Baumler 1997). The subspecies enterica is the most frequently found in food samples and is therefore the most relevant for human illnesses (Bertrand 2010). S. bongori and the other subspecies of S. enterica are rarely found in salmonellosis cases since they are associated with cold-blood invertebrates (Bertrand et al. 2008).
This paper reports on the development and validation of a Salmonella sp. detection system based on three detection levels. The first level detects all Salmonella spp., the second level detects all S. enterica and the third level detects all the Salmonella enterica enterica. Each level is composed of two to three SYBR®Green qPCR assays. The seven qPCR assays forming this detection system were developed to be run simultaneously on a single 96-well plate. Finally, this Combinatory SYBR®Green qPCR System (CoSYPS) is compared with commercial single-gene target Salmonella detection kits.
Material and methods
Bacterial strains
The bacterial strains used in this study are listed in Table 1. A panel of 111 bacterial strains (72 Salmonella strains and 39 other genera), two mould strains, two yeast strains and two viruses have been tested. These strains were obtained from National Reference Centres and Laboratories.
Bacterial growth conditions, DNA extraction and DNA quantification
Overnight cultures of each bacterial strain were grown in Brain-Heart Infusion (BHI) broth or Bolton broth (for Campylobacter) at the adequate temperature and oxygen condition. The total DNA from bacterial strains was extracted using the DNeasy Blood and Tissue Kit (Qiagen). Genomic DNA (gDNA) from yeast was extracted using the Invisorb Spin Plant Mini Kit (STRATEC Molecular GmbH). Fungal gDNA was extracted with ZR Fungal/Bacterial gDNA extraction (Zymo Research). Viral RNA was extracted using the RNeasy mini kit (Qiagen), and cDNA was obtained by reverse transcription using the Transcriptor high fidelity cDNA synthesis kit (Roche). The cDNA amplificability was checked with a specific PCR amplification. All kits were used according to the manufacturer's recommendations. The DNA quality was controlled on agarose gel, and the DNA concentration was measured using a Nanodrop® 2000 device according to the manufacturer's recommendations.
Development and in silico assessment of primer pairs
A uniform primer design approach was applied in the development of all primer pairs. The first step consisted of identifying genes of interest, either genus or species specific, by means of a bibliographic study (Chen et al. 1997; Galan et al. 1992; Gonzalez-Escalona et al. 2009; Parkhill et al. 2001; Porwollik et al. 2002; Soyer et al. 2009). The second step included the collection of DNA sequences relevant for the chosen targets retrieved from the NCBI public database (http://www.ncbi.nlm.nih.gov/sites/entrez). The primer pairs were designed, preferentially within conserved regions, using the “Primer 3” program (http://frodo.wi.mit.edu/primer3/ (Rozen and Skaletsky 2000) with the “product size range” specification set at “60 to 120 bp” and “primer size” optimal set at “22 bases”. An in silico test of the primer pairs was then performed. It consisted in a bioinformatic analysis carried out with the “wprimersearch” software available on wEMBOSS Open Source Software package (https://wemboss.uio.no/wEMBOSS/) (Rice et al. 2000; Sarachu and Colet 2005). This software mimics the PCR amplification of the tested primers on a database of bacterial genomes sequences from NCBI of 217 bacteria, representing 103 species belonging to 61 genera. When mismatches between primer and one of its targets were observed, degenerate nucleotides were introduced into the primer sequence. However, primer pairs with no degenerate nucleotides were always preferred. Only primer pairs that gave the expected in silico amplification were retained for the following steps.
Qualitative SYBR®Green qPCR assay and optimal primer concentration
All qPCR assay reactions were performed according to the protocol described in Barbau-Piednoir et al. (2013). For the interpretation of a SYBR®Green qPCR assay, two criteria were taken into consideration: the quantification cycle (Cq) value and the melting temperature of the amplicon (Tm). The Cq value represents the fractional cycle at which the PCR amplification reaches the threshold level for the reaction (Bustin 2000). Since it is a screening assay, a qualitative respond is required. To be considered as positive, a signal generated in SYBR®Green qPCR analysis should display an (exponential) amplification above the threshold level, with a single peak upon melting analysis giving a unique Tm value. A signal was considered as negative when no Cq value was obtained. The optimal concentration of the selected primer pairs was determined by testing different concentrations of each primer, between 250 and 1,000 nM. The concentrations giving the lowest Cq value without formation of a high level of primer dimer were selected. When using a positive sample at a concentration around the limit of detection (LOD), the primer dimer dissociation peak should not be higher than the positive sample dissociation peak. The primer pairs and their concentrations used in this study are presented in Table 2.
Selectivity test and accuracy calculation
Primer pairs selected from the in silico evaluation were tested in situ. In brief, the selectivity of the SYBR®Green qPCR assay is tested according to the ISO 22118 norm (2011) on target and non-target strains. The selectivity of an assay is assessed by inclusivity (target strains giving a positive signal) and exclusivity (non-target strains not giving a positive signal). The selectivity test is fully described in Barbau-Piednoir et al. (2013). From the selectivity test, the accuracy can be calculated. The accuracy represents the closeness of agreement between a test result and the accepted reference value (ISO 3534–1 1993). Its formula is found in ISO 16140 norm (2003).
Amplicon cloning and sequencing
In order to check the correctness of the amplicon obtained with each developed primer pair, the amplicons obtained by the amplification of S. enterica enterica Enteritidis (H.V.6.32) were cloned, sequenced and verified using the same approach as recently described by Barbau-Piednoir et al. (2013). Sequencing reactions were run on an ABI 3130xl at the Platform Biotechnology and Molecular Biology at the Scientific Institute of Public Health.
Full genome sequencing
The total DNA from S. enterica enterica serovar Namur was extracted with Blood & Cell Culture DNA Midi kit (Qiagen). The quality and quantity of the DNA were controlled with the 260/280-nm ratio and by spectrophotometer measurement, respectively. This DNA was sent to Baseclear for sequencing using the Illumina technology.
Dynamic range and calculation of the PCR efficiency
The dynamic range of a qPCR assay is the range of concentrations where it performs in a linear manner. It was assessed by the analysis in duplicate of a serial dilution in a carrier DNA (4 ng/μL Calf Thymus DNA (CTD) (Invitrogen) of pure strain DNA (1,000 to 0.01 theoretical genomic copies) of S. enterica enterica Enteritidis (H.V.6.32) and S. enterica enterica Typhimurium (H.II.32.32). The carrier DNA avoids the improper dilution due to low concentration of gDNA. This analysis allows the assessment of the coefficient of determination (R 2) and the PCR efficiency (E) for each SYBR®Green qPCR assays. The coefficient of determination (R 2) is an indicator of the correlation of data regarding the linear regression curve. The R 2 of the dynamic range curve should be above 0.98 (European Network of GMO Laboratories 2008). The PCR efficiency (E) should be between 89.6 and 110.2 % and can be calculated according to the formula reported by Rutledge and Cote (2003).
Sensitivity test
Primer pairs passing the selectivity test were subsequently examined for their sensitivity. Using serial dilution, the LOD of each SYBR®Green qPCR assays was evaluated. The LOD is defined as the lowest concentration of an analyte giving a positive result with a probability of 95 % (European Network of GMO Laboratories 2008). The strains used were S. enterica enterica Enteritidis (H.V.6.32) and S. enterica enterica Typhimurium (H.II.32.32). The calculation of the target genomic copy numbers for each dilution point was performed according to Barbau-Piednoir et al. (2013), considering the gDNA size of S. enterica enterica Enteritidis and Typhimurium as 4,685,848 and 4,857,432 bp (Acc. Num.: AM933172 and AE006468, respectively). To determine the LOD, a range of copy number between 10 and 0.1 theoretical copies was tested (i.e. 10, 5, 2, 1, 0.5, 0.2 and 0.1). Each dilution was tested in six replicates per plate, for both strains. Moreover, the analysis was performed three independent times, under repeatable conditions, resulting in 36 repeats for each dilution point. It has to be noticed that the dilution points beyond the theoretical single copy were carried out to assess the dilution series correctness. Indeed, as it is statistically impossible to get amplification in all replicates with the dilution points beyond one theoretical copy, none of these dilution points should give 100 % of positive signals.
Repeatability calculation
To evaluate the repeatability of the assay, independent tests were performed with the same protocol, with the same samples, by the same operator using the same apparatus within a short interval of time (ISO 3534–1 norm 1993). The repeatability limit (r) and the relative standard deviation of repeatability (RSDr) were calculated according to ISO 16140 norm (2003). RSDr should be ≤ 25 % for all the dilution range above the LOD (European Network of GMO Laboratories 2008). The RSDr and r values of the Cq values were calculated at each dilution point while the RSDr and r values of the Tm values were calculated with all the Tm values coupled with amplification (Cq <40).
Reproducibility study and calculation
To evaluate the reproducibility of the assays (ISO 3534–1 norm 1993), independent tests were performed with the same protocol, using the same samples, in two different laboratories, by two different operators using different apparatus (Bio-Rad iQ5 and ABI 7300). The nine tested samples were all gDNA from S. enterica enterica strains at different concentrations between 20 and 100 genomic copies per reaction. Each sample was analyzed in duplicate by each operator.
Two reproducibility measures can be calculated from these results: the relative standard deviation of reproducibility (RSDR) and the expanded uncertainty (U). RSDR represents the absolute value of the coefficient of variation. It is expressed in percent and is obtained by multiplying the reproducibility standard deviation by 100 and dividing this product by the reproducibility median (ISO 16140 2003). The RSDR should be ≤35 % for all the tested samples (European Network of GMO Laboratories 2008). The RSDR of the Cq values are calculated for the nine tested samples. The RSDR of the Tm values are calculated with all the Tm values coupled with an amplification (Cq <40).
The uncertainty is “the parameter associated with the result of a measurement that characterizes the dispersion of the values that could reasonably be attributed to the measurand”. The uncertainty can be expressed by the expanded uncertainty (U) which is the quantity defining “an interval about the result of a measurement that may be expected to encompass a large fraction of the distribution of values that could reasonably be attributed to the measurand”. The expanded uncertainty (U) is obtained by multiplying the combined standard uncertainty by a coverage factor (Joint Committee for Guides in Metrology 2008).
CoSYPS Salmonella
The seven qPCR SYBR®Green assays have been run, on the same plate with the same PCR program (Barbau-Piednoir et al. 2013) using the appropriate concentration of each primer (Table 2) on 104 copies of five gDNA extraction from pure culture of S. enterica enterica Enteritidis (H.VI.6.32), Salmonella enterica salamae (H.VII.72.10), Salmonella enterica arizonae (H.VI.61.2), Salmonella enterica diarizonae (NH.IV.4.1) and S. bongori (1224/72).
Comparison of CoSYPS Salmonella with other commercial kits
The CoSYPS Salmonella, composed of the seven SYBR®Green qPCR assays as detection/identification system, was compared with three Salmonella detection kits: iQ-Check™ Salmonella II from Bio-Rad, foodproof® Salmonella Detection Kit from Biotecon and GeneDisc® Salmonella spp. from Pall® Life Sciences (according to manufacturer's protocol). Eleven gDNA Salmonella samples from pure culture at 104 copies per assay have been tested in duplicate with the three detection systems.
Results
In silico selection of the primer pairs and optimisation of primer concentration
As a first step, specific genes of Salmonella genus, S. enterica species and S. enterica enterica subspecies were identified. For the specific detection of the Salmonella genus, invA and rpoD were retained. The rpoD gene encodes the RNA polymerase sigma 70 factor (Bai et al. 2012), while invA plays an important role during invasion of the intestinal cells (Galan et al. 1992). InvA is located in the Salmonella Pathogenicity Island I (SPI-I) (Baumler et al. 1997) shown to be present in all phylogenetic lineages of the Salmonella genus (Li et al. 1995). For the specific detection of the S. enterica, the chosen gene was iroB, which belongs to the Fur regulon that mediates gene regulation in response to iron availability (Ernst et al. 1978). This gene is not only present in S. enterica but also in all its subspecies (Porwollik et al. 2002). Finally, for the specific detection of S. enterica subsp. enterica, the safC and ratB genes were selected (Porwollik et al. 2002). SafC belongs to the safABCD operon (Folkesson et al. 2002) located in the Salmonella Pathogenicity Island 6 (SPI-6) (Parkhill et al. 2001). This operon encodes the Salmonella atypical fimbriae (Saf) pilus (Salih et al. 2008). RatB encodes an outer membrane protein essential in the colonization of the caecum by Salmonella.
During in silico evaluation of these primer pairs, some nucleotides were degenerated when necessary. Twenty primer pairs were first tested in situ with the “light” selectivity test (data not shown). From these, seven primer pairs were retained: invA-5 and rpoD-20 for the Salmonella spp. specific detection, iroB-192, iroB-2 and iroB-12 for the S. enterica specific detection and safC-1 and safC-13 for the S. enterica enterica detection. The optimal concentration of the seven primer was also evaluated (Table 2). These assays were then evaluated in vivo for their selectivity.
In situ optimisation of primer pair and targets
During in situ selectivity test, the primer pairs invA-5 and iroB-12 were found to less efficiently (latter Cq value) amplify S. bongori and Salmonella enterica houtenae, respectively, while strains S. enterica enterica Senftenberg, Namur and Krefeld were not amplified by the safC assays.
Optimisation of invA-5 and iroB-12 primer pairs
The locus containing the primer pair invA-5 in S. bongori and iroB-12 in S. enterica houtenae was sequenced (data not shown) and aligned with the published sequences in order to identify mismatches. The new degenerated primers invA-5-deg-F and iroB-12-deg-F are displayed in Table 2. The new assays performed with these degenerated primers amplified S. bongori and S. enterica houtenae, respectively, as efficiently (giving a comparable Cq value at the same concentration) as the other positive strains.
Optimisation of S. enterica enterica target
SafC could not be found in the genomes of both S. enterica enterica Senftenberg (CAGQ00000000.1 and AFCU00000000.1) and Namur (AWGG00000000) (see below). However, a short region (56 bp) into the STM0296 gene of S. enterica enterica Typhimurium (AE006468), upstream of the saf operon, was found to be highly conserved in S. enterica subsp. enterica including Senftenberg and Namur. Therefore, a new primer pair, named STM0296, was designed to amplify this 56-bp sequence (Table 2).
Determination of SYBR®Green qPCR assay selectivity
The primer pairs invA-5 and rpoD-20 amplified 100 % (72/72) of the Salmonella spp. tested strains, none (0/45) of the non-target strains nor the no template control (NTC) (Table 1). The iroB-192 and iroB-12 assays for S. enterica detection gave a specific amplification with 100 % (71/71) of S. enterica tested strains and 0 % (0/46) of the non-target strains and NTC (Table 1). Thus, these four assays were considered 100 % accurate for their target (S. enterica) giving 0 % of false positive and 0 % of false negative.
The four safC assays amplified 94.6 % (52/55) of the tested serovars and 0 % (0/61) of the non-target strains and NTC (Table 1). Primer pairs safC-10 and safC-13 were selected among four candidates since they amplified different loci of safC and gave the smallest amplicon size with two distinct Tm values (Table 2). These two assays give 0 % of false positive and 5.45 % (3/55) of false negative (serovars Senftenberg, Namur and Krefeld). The accuracy of these two assays is 97.4 % for detection of S. enterica enterica
The STM0296 assay amplifies 90.9 % (50/55) of the tested serovars and 0 % (0/61) of non-target strains and NTC (Table 1). This assay gives 0 % of false positive and 9.1 % (5/55) of false negative (serovars Heidelberg, Virchow, Perth, Hofit and Djakarta. The accuracy of this assay is 95.7 % for detection of S. enterica enterica. However, 100 % (55/55) of the tested serovars were amplified by at least one S. enterica enterica assay. Then, the combination of safC and STM0296 assays forming the level detection system was 100 % accurate for the specific detection of S. enterica enterica.
These seven assays tested on S. enterica enterica Enteritidis showed a unique band at the expected size upon agarose gel analysis (data not shown). The sequence of each amplicon was determined and shown to correspond to the sequence of S. enterica enterica Enteritidis (data not shown). In addition, the seven detection assays gave a unique melting peak with a specific melting temperature for each assay (Table 2).
SafC presence in S. enterica enterica Namur, Krefeld and Senftenberg
As indicated above, the genome of S. enterica enterica Namur was fully sequenced (data not shown). The presence of the Salmonella atypical fimbrial (saf) operon was neither been found in the Senftenberg strain whole genome shotgun contigs (CAGQ00000000.1 and AFCU00000000.1), nor in the Namur strain sequence (AWGG00000000). The S. enterica enterica Senftenberg, Namur and Krefeld strains were also PCR-tested using primers flanking, or located inside, the saf operon. None of them was positive confirming the absence of this operon in these three strains (data not shown). The lack of this saf operon is in accordance with the phylogenetic study of den Bakker et al. (2011) demonstrating the diversification of virulence factors content, mainly fimbrial operons, between two clades of S. enterica enterica.
Determination of SYBR®Green qPCR assay dynamic range and PCR efficiency
The seven developed assays performed in a linear manner between 1 and 1,000 copies as their R 2 values were between 0.982 and 0.999, above the required 0.98 (ENGL 2008) (Table 3). From the dynamic range analyses, the PCR efficiency (E) of each assay was calculated. The seven assays displayed PCR efficiencies ranging between 98 and 106 %, falling into the accepted limits of 89.6–110.2 % (ENGL 2008) (Table 3).
Determination of SYBR®Green qPCR assay sensitivity and repeatability
The LOD of the seven assays fell between two and ten copies (Table S1) complying with the requirement “between 1 and 10 CFU” (ISO 22118 norm 2011). The r values at the LOD of the Cq values ranged between 1.9 and 4.7 Cq and those of the Tm values ranged between 0.5 and 1 °C (Table 3). The RSDr values at LOD of the Ct values of the seven assays were between 1.7 and 5.1 % while those of the Tm values ranged between 0.2 and 0.5 % (Table 3). All these RSDr values were below ≤25 % as requested by the ENGL guidelines (ENGL 2008).
Determination of SYBR®Green qPCR assay reproducibility
For all the developed SYBR®Green qPCR assays, the RSDR values were below ≤35 % as requested by the ENGL guideline (ENGL 2008): between 0 and 1.01 % for the Tm values and between 0.00 and 7.34 % for the Ct values (Table S2). The expanded uncertainty at 99 % of confidence was also calculated from the reproducibility data from the eight samples. U was ranging between 0.39 and 1.47 °C for the Tm values and between 0.92 and 4.78 for the Cq values (Table S2).
CoSYPS Salmonella
The seven SYBR®Green qPCR assays were run together on the same plate. S. enterica enterica Enteritidis was amplified by the seven assays, whereas S. enterica arizonae, S. enterica salamae and S. enterica diarizonae subspecies were amplified by the four assays amplifying all the S. enterica subspecies. The S. bongori species was only amplified by the two assays that are amplifying all the species of the Salmonella genus (Table 4).
Comparison with other Salmonella qPCR detection kits
The CoSYPS Path Food detection/identification system based on seven qPCR assays allowed the detection of the eleven tested samples. The iQ-Check™ Salmonella II did not detect S. enterica enterica Senftenberg, isolated from a Chinese outbreaks that do not contain the SPI-1 (Hu et al. 2008) and the GeneDisc® Salmonella spp. did not detect S. bongori. The foodproof® of Biotecon detects all tested strains (Table 5).
Discussion
In order to significantly reduce the incidence of foodborne outbreaks, food chain surveillance is necessary (Official journal of the European Union 2004). Although standard protocols based on microbiology to detect foodborne pathogens are available (ISO 11290–1 norm 1996; ISO 16654 norm 2001; ISO 6579 norm 2002; ISO 10272 norm 2006), there are time consuming, labour intensive and target only single pathogenic bacteria. On the contrary, real-time PCR is much more rapid to detect foodborne pathogens (Postollec et al. 2011), and several assays have already been developed and published (e.g. Amagliani et al. 2010; Chen et al. 2000; Fukushima et al. 2009; Garrido et al. 2012a; Hein et al. 2006; Lund et al. 2004; McCabe et al. 2011; Perelle et al. 2004; Sharma 2002; Sharma and an-Nystrom 2003). Unfortunately, these assays are not harmonized: they are using different chemistry (SYBR®green or TaqMan®), different protocols and different validation procedures, hampering their use as coherent detection system. Moreover, these assays target a single DNA sequence, often virulence or associated virulence genes, potentially leading to false negative signal when the target is mutated or absent. Such drawbacks could be avoided in this study. For instance, the primers for invA and iroB were successfully degenerated to better amplify S. bongori and S. enterica houtenae, respectively, illustrating the variability of target genes. Also, comparison of different Salmonella qPCR detection kits demonstrated that single target kits were unable to detect S. bongori or an atypical S. enterica enterica Senftenberg lacking the entire Pathogenicity Island I (Hu et al. 2008) (Table 5). Therefore, strategy targeting several genes is highly advisable for the appropriate and efficient detection of pathogenic bacteria.
In this study, seven simplex qualitative detection SYBR®Green qPCR assays, targeting all Salmonella spp., one of the most important foodborne pathogen (European Food Safety Authority, European Centre for Disease Prevention and Control 2013), were successfully developed and in-house validated. Considering that all Salmonella spp. are pathogenic for human, these assays were developed targeting five distinct loci (invA, rpoD, iroB, safC and STM0296) for a full coverage of all the Salmonella species, subspecies and serovars. These assays were developed and validated, to be run simultaneously with the same PCR program, according a harmonized standard procedure described in Barbau-Piednoir et al. (2013).
The seven assays allowed a three-level detection system for the discrimination between the different species and subspecies of Salmonella (Table 4). Two assays allow the detection of the Salmonella genus bacteria based on invA and rpoD detection. Two assays permit the specific identification of the species S. enterica based on iroB detection. A third level is composed of two assays based on safC and an additional one targeting the STM0296 sequence. The last assay was developed to detect S. enterica enterica serovars presenting a deletion of safC. This deletion seems to be relatively frequent: present in three serovars (Senftenberg, Krefeld and Namur) among the 55 tested. This type of operon deletion was also reported in den Bakker et al. (2011) and hypothetically explained by a selective pressure. The three assays (safC-10, safC-13 and STM0296) specifically and exhaustively detected S. enterica subsp. enterica (Table 4), amplifying all the serovars tested in this study (Table 1).
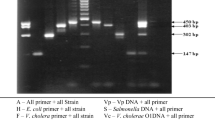
The SYBR®Green chemistry was preferred to the commonly used TaqMan® chemistry since SYBR®Green assays present the advantage of allowing the use of only two primers in the conserved region surrounding the more polymorphic sequence. As shown in Fig. 1, the design of a TaqMan® probe between invA-5 primers would have been impossible as polymorphisms are observed between all the Salmonella species and subspecies. In addition, SYBR®Green assays are cheaper and permit post-amplification verification of the amplicon specificity by melting temperature analysis (Postollec et al. 2011).
The seven SYBR®Green qPCR assays for Salmonella detection developed and validated in this paper could be combined with the four assays for Listeria detection previously described (Barbau-Piednoir et al. 2013) and run all together on a single plate. Since this approach is modular, detection assays can be added to detect a wider range of foodborne pathogens or emerging pathogens. The only requirement is to develop an assay able to be run with the same conditions, allowing its use in high throughput on the same 96-well plate.
Combined with a decision support system (Van den Bulcke et al. 2008; 2009), such detection method represents a very useful approach in managing the experimental analysis of samples for a high-quality screening system and a remarkable food surveillance tool which can be modulated in response to the laboratory needs. It will also considerably reduce the time and the cost of a sample analysis. Such simultaneous detection may be extremely useful when a global screening and rapid identification of the foodborne pathogens is requested, as in the case of a bio-emergency or outbreaks of unknown origin.
References
Amagliani G, Omiccioli E, Brandi G, Bruce IJ, Magnani M (2010) A multiplex magnetic capture hybridization and multiplex real-time PCR protocol for pathogen detection in seafood. Food Microbiol 27(5):580–585
Bai H, You Y, Yan H, Meng J, Xue X, Hou Z, Zhou Y, Ma X, Sang G, Luo X (2012) Antisense inhibition of gene expression and growth in gram-negative bacteria by cell-penetrating peptide conjugates of peptide nucleic acids targeted to rpoD gene. Biomaterials 33(2):659–667
Barbau-Piednoir E, Botteldoorn N, Yde M, Mahillon J, Roosens NH (2013) Development and validation of qualitative SYBR®Green real-time PCR for detection and discrimination of Listeria spp. and Listeria monocytogenes. Appl Microbiol Biotechnol 97(9):4021–4037
Baumler AJ (1997) The record of horizontal gene transfer in Salmonella. Trends Microbiol 5(8):318–322
Baumler AJ, Tsolis RM, Valentine PJ, Ficht TA, Heffron F (1997) Synergistic effect of mutations in invA and lpfC on the ability of Salmonella typhimurium to cause murine typhoid. Infect Immun 65(6):2254–2259
Bertrand S (2010) Rapport Annuel 2009 - Centre National de Référence des Salmonella et Shigella - Souches de Salmonella et Shigella isolées en Belgique en 2009
Bertrand S, Rimhanen-Finne R, Weill FX, Rabsch W, Thornton L, Perevoscikovs J, van Pelt W, Heck M (2008) Salmonella infections associated with reptiles: the current situation in Europe. Euro Surveill, 13(24)
Beutin L, Jahn S, Fach P (2009) Evaluation of the 'GeneDisc' real-time PCR system for detection of enterohaemorrhagic Escherichia coli (EHEC) O26, O103, O111, O145 and O157 strains according to their virulence markers and their O- and H-antigen-associated genes. J Appl Microbiol 106(4):1122–1132
Botteldoorn N, Heyndrickx M, Rijpens N, Herman L (2003) Detection and characterization of verotoxigenic Escherichia coli by a VTEC/EHEC multiplex PCR in porcine faeces and pig carcass swabs. Res Microbiol 154(2):97–104
Botteldoorn N, Van CE, Grijspeerdt K, Werbrouck H, Haesebrouck F, Donne E, D'Haese E, Heyndrickx M, Pasmans F, Herman L (2006) Real-time reverse transcription PCR for the quantification of the mntH expression of Salmonella enterica as a function of growth phase and phagosome-like conditions. J Microbiol Methods 66(1):125–135
Bustin SA (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 25(2):169–193
Chen S, Yee A, Griffiths M, Larkin C, Yamashiro CT, Behari R, Paszko-Kolva C, Rahn K, De Grandis SA (1997) The evaluation of a fluorogenic polymerase chain reaction assay for the detection of Salmonella species in food commodities. Int J Food Microbiol 35(3):239–250
Chen W, Martinez G, Mulchandani A (2000) Molecular beacons: a real-time polymerase chain reaction assay for detecting Salmonella. Anal Biochem 280(1):166–172
den Bakker HC, Moreno Switt AI, Govoni G, Cummings CA, Ranieri ML, Degoricija L, Hoelzer K, Rodriguez-Rivera LD, Brown S, Bolchacova E, Furtado MR, Wiedmann M (2011) Genome sequencing reveals diversification of virulence factor content and possible host adaptation in distinct subpopulations of Salmonella enterica. BMC Genomics 12:425
Ernst JF, Bennett RL, Rothfield LI (1978) Constitutive expression of the iron-enterochelin and ferrichrome uptake systems in a mutant strain of Salmonella typhimurium. J Bacteriol 135(3):928–934
European Commission (2011) SANCO/13004/2011: Lessons learned from the 2011 outbreak of Shiga toxin-producing Escherichia coli (STEC) O104:H4 in sprouted seeds, p 23
European Commission (2012) The Rapid Alert System for food and feed (RASFF): Annual report 2011. doi:10.2772/96194
European Food Safety Authority, European Centre for Disease Prevention and Control (2013) The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-borne Outbreaks in 2011. EFSA J 11(4):3129. doi:10.2903/j.efsa.2013.3129
European Network of GMO Laboratories (2008) Definition of minimum performance requirements for analytical methods of GMO testing, p 8. European Commission, Brussels
Folkesson A, Lofdahl S, Normark S (2002) The Salmonella enterica subspecies I specific centisome 7 genomic island encodes novel protein families present in bacteria living in close contact with eukaryotic cells. Res Microbiol 153(8):537–545
Fukushima H, Katsube K, Tsunomori Y, Kishi R, Atsuta J, Akiba Y (2009) Comprehensive and rapid real-time PCR analysis of 21 foodborne outbreaks. Int J Microbiol 2009:917623
Galan JE, Ginocchio C, Costeas P (1992) Molecular and functional characterization of the Salmonella invasion gene invA: homology of InvA to members of a new protein family. J Bacteriol 174(13):4338–4349
Garrido A, Chapela MJ, Roman B, Fajardo P, Lago J, Vieties JM, Cabado AG (2012a) A new multiplex real-time PCR developped method for Salmonella spp. and Listeria monocytogenes detection in food and environmental samples. Food Control 30(1):76–85
Garrido A, Chapela MJ, Roman B, Ferreira M, Lago J, Vieites JM, Cabado AG (2012b) Development of a multiplex real-time PCR method for simultaneous detection of Salmonella enterica, Shigella flexneri and Listeria monocytogenes in processed food samples. Eur Food Res Technol 234(4):571–580
Gomez-Aldapa CA, del Refugio Torres-Vitela M, Villarruel-Lopez A, Castro-Rosas J (2012) The role of foods in Salmonella infection. In: Salmonella - A Dangerous Foodborne Pathogen. InTech, Rijeka, pp 21–46, doi:10.5772/28316
Gonzalez-Escalona N, Hammack TS, Russell M, Jacobson AP, De Jesus AJ, Brown EW, Lampel KA (2009) Detection of live Salmonella sp. cells in produce by a TaqMan-based quantitative reverse transcriptase real-time PCR targeting invA mRNA. Appl Environ Microbiol 75(11):3714–3720
Guibourdenche M, Roggentin P, Mikoleit M, Fields PI, Bockemühl J, Grimont PAD, Weill FX (2010) Supplement 2003–2007 (No. 47) to the White-Kauffmann-Le Minor scheme. Res Microbiol 161(1):26–29
Hein I, Flekna G, Krassnig M, Wagner M (2006) Real-time PCR for the detection of Salmonella spp. in food: an alternative approach to a conventional PCR system suggested by the FOOD-PCR project. J Microbiol Methods 66(3):538–547
Hu Q, Coburn B, Deng W, Li Y, Shi X, Lan Q, Wang B, Coombes BK, Finlay BB (2008) Salmonella enterica serovar Senftenberg human clinical isolates lacking SPI-1. J Clin Microbiol 46(4):1330–1336
International Organization for Standardization (ISO) (1993) ISO 3534–1:1993 Statistics-Vocabulary and Symbols—Part 1. ISO, Geneva
International Organization for Standardization (ISO) (1996) ISO 11290–1:1996 Microbiologie des aliments - Méthode horizontale pour la recherche et le dénombrement de Listeria monocytogenes - Partie 1: Méthode de recherche. ISO, Geneva
International Organization for Standardization (ISO) (2001) ISO 16654:2001 Microbiologie des aliments - Méthode horizontale pour la recherche des Escherichia coli O157. ISO, Geneva
International Organization for Standardization (ISO) (2002) ISO 6579:2002: Microbiologie des aliments - Méthode horizontale pour la recherche des Salmonella spp. ISO, Geneva
International Organization for Standardization (ISO) (2003) ISO 16140:2003: Microbiologie des aliments - Protocole pour la validation des méthodes alternatives. ISO, Geneva
International Organization for Standardization (ISO) (2006) ISO 10272–1:2006 Microbiologie des aliments - Méthode horizontale pour la recherche et le dénombrement de Campylobacter spp. - Partie 1: Méthode de recherche. ISO, Geneva
International Organization for Standardization (ISO) (2011) ISO 22118:2011-Microbiology of food and animal feeding stuffs - Polymerase chain reaction (PCR) for the detection and quantification of food-borne pathogens - Performance characteristics of molecular detection methods. ISO, Geneva
Joint Committee for Guides in Metrology (2008) Evaluation of measurement data - Guide to the expression of uncertainty in measurement. Joint Committee for Guides in Metrology, Paris
Kim HJ, Cho JC (2010) Simple and rapid detection of Listeria monocytogenes in fruit juice by real-time PCR without enrichment culture. Food Control 21(10):1419–1423
Li J, Ochman H, Groisman EA, Boyd EF, Solomon F, Nelson K, Selander RK (1995) Relationship between evolutionary rate and cellular location among the Inv/Spa invasion proteins of Salmonella enterica. Proc Natl Acad Sci U S A 92(16):7252–7256
Löfström C, Hansen F, Hoorfar J (2010) Validation of a 20-h real-time PCR method for screening of Salmonella in poultry faecal samples. Vet Microbiol 144(3–4):511–514
Lund M, Nordentoft S, Pedersen K, Madsen M (2004) Detection of Campylobacter spp. in chicken fecal samples by real-time PCR. J Clin Microbiol 42(11):5125–5132
McCabe EM, Burgess CM, O'Regan E, McGuinness S, Barry T, Fanning S, Duffy G (2011) Development and evaluation of DNA and RNA real-time assays for food analysis using the hilA gene of Salmonella enterica subspecies enterica. Food Microbiol 28(3):447–456
Medus C, Smith KE, Bender JB, Besser JM, Hedberg CW (2006) Salmonella outbreaks in restaurants in Minnesota, 1995 through 2003: evaluation of the role of infected foodworkers. J Food Prot 69(8):1870–1878
Murphy NM, McLauchlin J, Ohai C, Grant KA (2007) Construction and evaluation of a microbiological positive process internal control for PCR-based examination of food samples for Listeria monocytogenes and Salmonella enterica. Int J Food Microbiol 120(1–2):110–119
Official Journal of the European Union (2004) Regulation (Ec) No 882/2004 of The European Parliament and of The Council Of 29 April 2004 on official controls performed to ensure the verification of compliance with feed and food law, animal health and animal welfare rules. p 59, European Union, Brussles
Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Churcher C, Mungall KL, Bentley SD, Holden MT, Sebaihia M, Baker S, Basham D, Brooks K, Chillingworth T, Connerton P, Cronin A, Davis P, Davies RM, Dowd L, White N, Farrar J, Feltwell T, Hamlin N, Haque A, Hien TT, Holroyd S, Jagels K, Krogh A, Larsen TS, Leather S, Moule S, O'Gaora P, Parry C, Quail M, Rutherford K, Simmonds M, Skelton J, Stevens K, Whitehead S, Barrell BG (2001) Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848–852
Perelle S, Josefsen M, Hoorfar J, Dilasser F, Grout J, Fach P (2004) A LightCycler real-time PCR hybridization probe assay for detecting food-borne thermophilic Campylobacter. Mol Cell Probes 18(5):321–327
Porwollik S, Wong RM, McClelland M (2002) Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc Natl Acad Sci U S A 99(13):8956–8961
Postollec F, Falentin H, Pavan S, Combrisson J, Sohier D (2011) Recent advances in quantitative PCR (qPCR) applications in food microbiology. Food Microbiol 28(5):848–861
Pui CF, Wong WC, Chai LC, Tunung R, Jeyalechumi P, Noor Hidayah MS, Ubong A, Farinazleen MG, Cheah YK, Son R (2011) Review article: Salmonella: a foodborne pathogen. Int Food Res J 18:465–473
Rice P, Longden I, Bleasby A (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16(6):276–277
Rozen S, Skaletsky H (2000) Primer 3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386
Rutledge RG, Cote C (2003) Mathematics of quantitative kinetic PCR and the application of standard curves. Nucleic Acids Res 31(16):e93
Salih O, Remaut H, Waksman G, Orlova EV (2008) Structural analysis of the Saf pilus by electron microscopy and image processing. J Mol Biol 379(1):174–187
Sarachu M, Colet M (2005) wEMBOSS: a web interface for EMBOSS. Bioinformatics 21(4):540–541
Scharff RL (2012) Economic burden from health losses due to foodborne illness in the United States. J Food Prot 75(1):123–131
Sharma VK (2002) Detection and quantitation of enterohemorrhagic Escherichia coli O157, O111, and O26 in beef and bovine feces by real-time polymerase chain reaction. J Food Prot 65(9):1371–1380
Sharma VK, an-Nystrom EA (2003) Detection of enterohemorrhagic Escherichia coli O157:H7 by using a multiplex real-time PCR assay for genes encoding intimin and Shiga toxins. Vet Microbiol 93(3):247–260
Singh J, Batish V, Grover S (2012) Simultaneous detection of Listeria monocytogenes and Salmonella spp. in dairy products using real time PCR-melt curve analysis. J Food Sci Technol 49(2):234–239
Soyer Y, Orsi RH, Rodriguez-Rivera LD, Sun Q, Wiedmann M (2009) Genome wide evolutionary analyses reveal serotype specific patterns of positive selection in selected Salmonella serotypes. BMC Evol Biol 9:264
Van den Bulcke M, Lievens A, Leunda-Casi A, Mbongolo Mbella G, Barbau-Piednoir E, Sneyers M (2008) Transgenic Plant Event Detection, WO/2008/092866 (patent)
Van den Bulcke M, Lievens A, Barbau-Piednoir E, Mbongolombella G, Roosens N, Sneyers M, Leunda-Casi A (2009) A theoretical introduction to "Combinatory SYBR®Green qPCR Screening", a matrix-based approach for the detection of materials derived from genetically modified plants. Anal Bioanal Chem 396(6):2113–2123
Acknowledgments
The authors would like to greatly thank all the laboratories that provided the strains and the companies Bio-Rad, Pall® Corporation and BIOTECON Diagnostics GmbH which agreed with the comparison test and freely provided the kits. This project was funded by an Ylieff grant from the Belgian Federal Public Service: Public Heath, Food Chain Safety and Environment. The authors would like to greatly thank Alexandra Duarte who performed the reproducibility analyses in the Food Pathogens Laboratory of the ISP-WIV.
Author information
Authors and Affiliations
Corresponding author
Additional information
Nancy H. Roosens and Nadine Botteldoorn contributed equally to this work
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 46 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Barbau-Piednoir, E., Bertrand, S., Mahillon, J. et al. SYBR®Green qPCR Salmonella detection system allowing discrimination at the genus, species and subspecies levels. Appl Microbiol Biotechnol 97, 9811–9824 (2013). https://doi.org/10.1007/s00253-013-5234-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-013-5234-x