Abstract
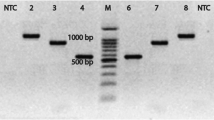
Effective monitoring of Salmonella contamination in seafood processing to conform the requirements of HACCP is a great challenge today. Such challenges can be effectively addressed, if the conventional detection methods are replaced with DNA-based molecular methods. Accordingly, it was aimed to develop a robust PCR protocol for specific detection of Salmonella spp. Out of the different primers screened, one pair of primers developed in this study targeting invA gene demonstrated 100% inclusivity for a wide range of Salmonella serotypes and 100% exclusivity for wide range of non-target species. The in silico analysis of the nucleotide sequence obtained from the PCR product suggests its potential as a hybridization probe for genus specific detection of Salmonella spp. contamination. The PCR protocol was sensitive enough to detect 15 cells per reaction using crude DNA prepared within a short time directly from artificially contaminated shrimp tissue. The study demonstrated that the result of PCR reaction can come out on the same day of sample arrival. Incorporation of this pair of primers in a multiplex PCR designed for simultaneous detection of four common seafood-borne human pathogens yielded 147 bp, 302 bp, 403 bp, and 450 bp distinct DNA bands specifically targeting E. coli, toxigenic Vibrio cholerae, Salmonella spp., and V. parahaemolyticus, respectively in a single PCR tube. The PCR methods developed in this study has the potential to be used in the seafood processing plants for effective monitoring of CCPs required for implementation of HACCP-based quality assurance system.

Similar content being viewed by others
References
USFDA (2011) FDA and EPA safety levels in regulations and guidance. In: Fish and fishery products hazards and controls guidance, fourth edition, appendix 5. FDA, CFSAN, pp. 439–442
Bailey JL (1998) Detection of Salmonella cells within 24 to 26 hours in poultry samples with polymerase chain reaction BAX system. J Food Prot 61(7):792–795
Hendriksen RS, Vieira AR, Karlsmose S, Fo L, Wong DM, Jensen AB, Wegener HC, Aarestrup FM (2011) Global monitoring of Salmonella serovar distribution from the WHO global foodborne infections network country data bank: results of quality assured laboratories from 2001 to 2007. Foodborne Pathog Dis 8(8):887–900
Choudhury M, Borah P, Sarma HK, Barkalita LM, Deka NK, Hussain I, Hussain M (2016) Multiplex-PCR assay for detection of some major virulence genes of Salmonella enterica serovars from diverse sources. Curr Sci (00113891), 111(7)
Popoff MY, Bockemühl J, Brenner FW (1998) Supplement (no. 42) to the Kauffmann-white scheme. Res Microbiol 2000151(1):63–65
Kwang J, Littledike ET, Keen JE (1996) Use of the polymerase chain reaction for Salmonella detection. Lett Appl Microbiol 22(1):46–51
Abu-Salem FM, Arab EA (2010) Chemical properties, microbiological quality and sensory evaluation of chicken and duck liver paste (foiegras). Grasas Aceites 61(2):126–135
Andrews WH, Hammack TS (2001) Salmonella. In: Bacteriological analytical manual online, official methods of analysis of AOAC international chapter 5. Center for food safety and applied nutrition, U.S.FDA (http://www.cfsan.fda.gov/~ebam/ bam-toc.Html)
Malorny B, Hoorfar J, Bunge C, Helmuth R (2003) Multicenter validation of the analytical accuracy of Salmonella PCR: towards an international standard. Appl Environ Microbiol 69(1):290–296
Ausubel FM, Brent R, Kingstone RE, Moore DD, Seidman JG, Smith JA, Struhl K (1987) Curr Protoc Mol Biol. John Wiley & Sons, Inc., USA
Rodriguez-Lazaro D, Gonzalez-García P, Delibato E, De Medici D, García-Gimeno RM, Valero A, Hernandez M (2014) Next day Salmonella spp. detection method based on real-time PCR for meat, dairy and vegetable food products. Int J Food Microbiol 184:113–120
Cocolin L, Manzano M, Cantoni C, Comi G (1998) Use of polymerase chain reaction and restriction enzyme analysis to directly detect and identify Salmonella Typhimurium in food. J Appl Microbiol 85(4):673–677
Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA (2007) Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res 35(suppl_2):W71–W74
Sambrook J, Russel DW (2001) Molecular cloning: a laboratory manual, 3rd ed., Vol. 1 and 2. Cold Spring Harbour Laboratory Press, Cold Spring Harbour, pp 1.1–14.53
Olsen JE (2000) DNA-based methods for detection of food-borne bacterial pathogens. Food Res Int 33(3):257–266
Tsen HY, Liou JW, Lin CK (1994) Possible use of a polymerase chain reaction method for specific detection of Salmonella in beef. J Ferment Bioeng 77(2):137–143
Fitts R, Diamond M, Hamilton C, Neri M (1983) DNA-DNA hybridization assay for detection of Salmonella spp. in foods. Appl Environ Microbiol 46(5):1146–1151
De la Cruz MA, Zhao W, Farenc C, Gimenez G, Raoult D, Cambillau C, Gorvel JP, Méresse SA (2013) Toxin-antitoxin module of Salmonella promotes virulence in mice. PLoS Pathog 9(12):e1003827. https://doi.org/10.1371/journal.ppat.1003827
Le ML, Popoff MY (1987) Designation of Salmonella enterica sp. nov., nom. Rev., as the type and only species of the genus Salmonella: request for an opinion. Int J Syst Evol Microbiol 37(4):465–468
Boyd EF, Wang FS, Whittam TS, Selander RK (1996) Molecular genetic relationships of the salmonellae. Appl Environ Microbiol 62(3):804–808
Fàbrega A, Vila J (2013) Salmonella enteric serovar typhimurium skills to succeed in the host: virulence and regulation. Clin Microbiol Rev 26(2):308–341
Chiu CH, Ou JT (1996) Rapid identification of Salmonella serovars in feces by specific detection of virulence genes, invA and spvC, by an enrichment broth culture-multiplex PCR combination assay. J Clin Microbiol 34(10):2619–2622
Pavlova B, Volf J, Ondrackova P, Matiasovic J, Stepanova H, Crhanova M, Karasova D, Faldyna M, Rychlik I (2011) SPI-1-encoded type III secretion system of Salmonella enterica is required for the suppression of porcine alveolar macrophage cytokine expression. Vet Res 42(1):16
Galan JE, Ginocchio C, Costeas P (1992) Molecular and functional characterization of the Salmonella invasion gene invA: homology of InvA to members of a new protein family. J Bacteriol 174:4338–4349
Rahn K, De Grandis SA, Clarke RC, McEwen SA, Galan JE, Ginocchio C, Curtiss RI, Gyles CL (1992) Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol Cell Probes 6(4):271
Umesha S, Manukumar HM, Raghava SA (2016) Rapid method for isolation of genomic DNA from food-borne fungal pathogens. 3 Biotech; 6(2)
Rossen L, Nørskov P, Holmstrøm K, Rasmussen OF (1992) Inhibition of PCR by components of food samples, microbial diagnostic assays and DNA-extraction solutions. Int J Food Microbiol 17(1):37–45
Widjojoatmodjo MN, Fluit AC, Verhoef J (1995) Molecular identification of bacteria by fluorescence-based PCR-single-strand conformation polymorphism analysis of the 16S rRNA gene. J Clin Microbiol 33(10):2601–2606
Roewer L (2013) DNA fingerprinting in forensics: past, present, future. Investig Genet.; 4
De Medici D, Croci L, Delibato E, Di Pasquale S, Filetici E, Toti L (2003) Evaluation of DNA extraction methods for use in combination with SYBR green I real-time PCR to detect Salmonella enterica serotype enteritidis in poultry. AEM 69(6):3456–3461
Mukhopadhyay A, Mukhopadhyay UK (2007) Novel multiplex PCR approaches for the simultaneous detection of human pathogens: Escherichia coli 0157: H7 and Listeria monocytogenes. J Microbiol Methods 68(1):193–200
Zhai L, Yu Q, Bie X, Lu Z, Lv F, Zhang C, Kong X, Zhao H (2014) Development of a PCR test system for specific detection of Salmonella Paratyphi B in foods. FEMS Microbiol Lett 355(1):83–89
Ogunremi D, Nadin-Davis S, Dupras AA, Márquez IG, Omidi K, Pope L, Devenish J, Burke T, Allain R, Leclair D (2017) Evaluation of a multiplex PCR assay for the identification of Salmonella serovars Enteritidis and Typhimurium using retail and Abattoir samples. J Food Prot 80(2):295–301
Yamasaki E, Sakamoto R, Matsumoto T, Maiti B, Okumura K, Morimatsu F, Balakrish Nair G, Kurazono H (2017) Detection of Cholera Toxin by an Immuno-chromatographic Test Strip. Microbial Toxins: Methods and Protocols :1–7
Eschbach E, Martin A, Huhn J, Seidel C, Heuer R, Schumacher JH, Ulrich S, Axe JO, Konietzny A, Strauch E, Oberheitmann B (2017) Detection of enteropathogenic Vibrio parahaemolyticus, Vibrio cholerae and Vibrio vulnificus: performance of real-time PCR kits in an interlaboratory study. Eur Food Res Technol :1–8
Li, R, Chiou J, Chan EW, Chen S (2016) A novel PCR-based approach for accurate identification of Vibrio parahaemolyticus. Front Microbiol.;7
Karus A, Ceciliani F, Bonastre AS, Karus V (2017) Development of simple multiplex real-time PCR assays for foodborne pathogens detection and identification on lightcycler. Mac Vet Rev 40(1):53–58
Shimada T, Arakawa E, Itoh K, Okitsu T, Matsushima A, Asai Y, Takeda Y (1994) Extended serotyping scheme for Vibrio cholerae. Curr Microbiol 28(3):175–178
Weynberg KD,Voolstra CR, Neave MJ, Buerger P, Van Oppen MJ (2015) From cholera to corals: viruses as drivers of virulence in a major coral bacterial pathogen. Sci Rep;5
Taniguchi H, Hirano H, Kubomura S, Higashi K, Mizuguchi Y (1986) Comparison of the nucleotide sequences of the genes for the thermostable direct hemolysin and the thermolabile hemolysin from Vibrio parahaemolyticus. Microb Pathog 1(5):425–432
Acknowledgements
The authors are highly grateful to Indian Council of Agricultural Research (ICAR), New Delhi, for providing fellowship to undertake the research program. The authors are also thankful to the Director, CIFE, Mumbai, India, for providing necessary facility to conduct the research program.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Luis Augusto Nero
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sahu, B., Singh, S.D., Behera, B.K. et al. Rapid detection of Salmonella contamination in seafoods using multiplex PCR. Braz J Microbiol 50, 807–816 (2019). https://doi.org/10.1007/s42770-019-00072-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-019-00072-8