Abstract
The Bacteriophage λ capsid protein gpD has been used extensively for fusion polypeptides that can be expressed from plasmids in Escherichia coli and remain soluble. In this study, a genetically controlled dual expression system for the display of enhanced green fluorescent protein (eGFP) was developed and characterized. Wild-type D protein (gpD) expression is encoded by λ Dam15 infecting phage particles, which can only produce a functional gpD protein when translated in amber suppressor strains of E. coli in the absence of complementing gpD from a plasmid. However, the isogenic suppressors vary dramatically in their ability to restore functional packaging to λDam15, imparting the first dimension of decorative control. In combination, the D-fusion protein, gpD::eGFP, was supplied in trans from a multicopy temperature-inducible expression plasmid, influencing D::eGFP expression and hence the availability of gpD::eGFP to complement for the Dam15 mutation and decorate viable phage progeny. Despite being the worst suppressor, maximal incorporation of gpD::eGFP into the λDam15 phage capsid was imparted by the SupD strain, conferring a gpDQ68S substitution, induced for plasmid expression of pD::eGFP. Differences in size, fluorescence and absolute protein decoration between phage preparations could be achieved by varying the temperature of and the suppressor host carrying the pD::eGFP plasmid. The effective preparation with these two variables provides a simple means by which to manage fusion decoration on the surface of phage λ.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The small-size and the enormous diversity of variants that can be fused to the bacteriophage capsid make bacteriophage ideal candidates for many applications across all industries including targeted therapy and detection in medicine to conjugation with macromolecules, plant science and nanoparticles in materials science (Willats 2002; Petty et al. 2007). Bacteriophage can also be made in mass quantities very quickly and at a relatively low-cost. The potential for phage as gene delivery vectors is strong since bacteriophage have many of the desirable properties of both the viral and non-viral systems with few of the drawbacks (Larocca et al. 2002; Seow and Wood 2009).
Phage display development has undergone considerable growth since Smith (1985) successfully displayed EcoRI endonuclease between the domains of pIII, the filamentous phage minor coat protein. Modern phage display practices with M13 will typically utilize the N-terminals of coat proteins pIII and pVIII (Kehoe and Kay 2005); though, proteins have been successfully displayed on all five major capsid proteins (Nilsson et al. 2000). While filamentous phage display systems have contributed greatly to the field, lytic based phage display systems, notably bacteriophages lambda (λ), T4, and T7, offer more flexible alternatives that can display both hydrophilic (Willats 2002; Vaccaro et al. 2006; Mikawa et al. 1996) and toxic fusion proteins (Kasman et al. 1998; Marzari et al. 1997; Vilchez and Jacoby 2004), previously limiting in filamentous phage systems (Nilsson et al. 2000; Ptashne 2004).
Fully formed, bacteriophage lambda has a linear dsDNA genome harbored within an icosahedral capsid comprised of major capsid proteins gpE and gpD that assemble the head in two main steps: prohead assembly followed by DNA packaging. Here, the assembled lambda prohead shell is composed mainly of gpE, in approximately 415 copies, and DNA packaging requires a conformational change to the prohead via the addition of gpD to the prohead (Georgopoulos et al. 1983; Yang et al. 2000). The addition of gpD then occurs in 405–420 trimer-clustered molecules (Beghetto and Gargano 2011) that act to increase the head volume and stability and is essential for the packaging for a full-length λ genome (Georgopoulos et al. 1983; Yang et al. 2000). In contrast, gpD-deficient viruses can package up to 82 % of the wild-type genome (Dokland and Murialdo 1993); although, they will have to be stabilized by magnesium ions and are extremely sensitive to EDTA (Yang et al. 2000; Sternberg et al. 1979).
Since initial λ phage display fusions to the gpV major tail (Dunn 1995), phage display fusions have since moved onto expression on gpD which confer a high decoration capacity per phage particle (up to 420), (Mikawa et al. 1996; Maruyama et al. 1994) and provide a selectable phenotype as a conditionally required packaging protein for full-length λ genome. The gpD protein assembles in trimers that are incorporated as prominent protrusions on the surface of the phage capsid making them more accessible for binding to external target molecules (Dokland and Murialdo 1993; Sternberg and Hoess 1995). Furthermore, gpD fusions of various sizes have been successfully fused to both the amino and carboxy termini of the protein suggesting that the display of the fusions does not jeopardize the function of the bacteriophage or prevent fusion proteins from binding the capsid (Vilchez and Jacoby 2004; Dokland and Murialdo 1993; Sternberg and Hoess 1995). The C-terminus is generally more tolerant of fusions since the N-terminus is located closer to the 3-fold axis of the gpD trimer (Yang et al. 2000) and may be involved with interacting with gpE. Upon displaying scFV antibodies to both the N and C Terminals of gpD N-terminal fusions were found to impart low recombinant protein loading compared to the C-terminal fusions (50 % compared to 88 %) (Petty et al. 2007).
Issues with λ display limit the efficiency and utility of this powerful system. In particular, capsid fusions often interfere with lambda phage morphogenesis, where a positive charge close to the signal sequence cleavage site or a large protein domain may impede capsid assembly (Beghetto and Gargano 2011). An approach to overcome this limitation is the use of dual expression systems. Originally developed for filamentous phage systems, dual expression systems in λ display first employed amber-suppression mediated control of gpD fusions (Mikawa et al. 1996). The original display system expressed gpD fusions to the N or the C termini in the presence of unfused gpD alleles varied by suppressor tRNA translation of Dam15 in the Escherichia coli host (Mikawa et al. 1996). Zanghi et al. (2005) later developed a plasmid-based dual expression system employing two independently selectable plasmid vectors for wild-type D and the D::fusion, expressed from gpD-λ lysates, finding that problematic protein fusions such as that of fibronectin type III, could now be overcome due to lower levels of interference (Zanghi et al. 2005). They further expanded this approach to incorporate fusions on both the gpD head and gpZ tail proteins, expressing both with high copy number (Zanghi et al. 2007). Another variation includes the use of a plasmid encoding a genomic copy of gpD with an amber mutation at the 5′ end of the gene and another copy of the gene under the control of an inducible promoter (Beghetto et al. 2001), where phage grown on amber suppressors incorporated gpD expressed from the genomic copy and the recombinant gpD from the inducible copy (Beghetto et al. 2001). And, most recently, a fusion gene was placed under the control of a temperature-sensitive promoter, allowing for repression by growing the bacterial cells at lower temperature, keeping the plasmid encoding the fusion gene repressed until the expression of the plasmid was needed, providing varied fusion decoration levels in resultant phage (Hayes et al. 2010).
While virtually limitless in application, lytic phage display does not come without limitations, particularly when considering the size and copy number of the displayed peptides as a result of the current display system design. The phage display system practice will need the optimization of fusion coat proteins to wild-type ones since a high ratio of fusion protein may lead to the inefficient assembly of phage particles and depending on the application a low ratio may not elicit the desired results (Gupta et al. 2003). In this work, we endeavored to construct and characterize a fine-tunable λ phage display system to control and predict the incorporation of gpD::fusions into the phage capsid.
Methods and materials
Strains and plasmids
Lambda phages, E. coli K-12 strains and plasmids used in this work are shown in Table 1. Amber suppressor strains (SupD, E, F) of W3101 were constructed in 2 steps. First, P1 rev6-mediated transduction of a tetracycline resistance (TcR) marker from CAG12077 to recipient amber suppressor (AS) strains DS-3, and W3899 was performed to link the AS marker to the TcR marker. K1227 (SupF) already possessed a linked TcR marker (see Table 1). P1 rev6 was used to cotransduce the AS and TcR marker (<2 min distance) into recipient W3101 cells, which were screened with λimm21Dam15 (λF7) and λSam7 phage to ensure transfer of the AS alleles, respectively linked to the TcR marker, derived from donor strains, to the W3101 non-suppressor (Sup−) recipient strain. The construction of plasmid pPL451-gpD::eGFP (herein referred to as pD::eGFP) was previously described (Sokolenko et al. 2012), where gpD::eGFP expression from this multicopy plasmid (pPL451) is governed by a temperature-sensitive allele of the λ CI857 repressor. The D::eGFP sequence designed creates a C-terminal eGFP translational fusion with λ D capsid gene, separated by an in-frame short linker encoding amino acids (TSGSGSGSGSGT) followed by a KpnI cut site to allow for removal and exchange of eGFP.
Plasmid pPL451-gpD (herein referred to as pD) was constructed by digesting pPL451-gpD::eGFP by KpnI removing all but the last 30 C-terminal bp of eGFP. Plasmid pPL451 was digested with HpaI and pPL451 gpD::eGFP and pPL451 gpD were double digested with HpaI and NcoI and the digestion pattern analyzed to ensure cloning accuracy.
Phage amplification and purification
Cultures of transformed W3101 Sup+/Sup− [pPL451gpD::eGFP] (pD::eGFP) E. coli cells were grown on plates at 30, 35, 37, or 39 °C overnight, while cultures of W3101 [pPL451gpD] (pD) were grown at 37 °C only, prior to the addition of primary lysate dilutions. Dilutions of primary lysates (1:10) were prepared in 1 mL of TN buffer (0.01 M Tris–HCl and 0.1 M NaCl, pH 7.8, (Fisher Scientific, USA). Lysate dilutions were added to 0.3 ml of cells, incubated for 2 hr at experimental temperature prior to adding 3 ml of top agar (Bacto Tryptone and Bacto Agar from Difco Laboratories, Sparks, MD) and plates were incubated overnight at the temperature that the experimental temperature. Plate lysates were then prepared by adding 10 mL of TN buffer to the surface of the plate, incubating for 8 hr at 4 °C, then transferring solution and top agar to a conical tube, mixing and centrifuging at 12 K RPM (Avanti J-E Centrifuge, Beckman Coulter, Mississauga Canada) at 4 °C for 20 min. The supernatant was poured into a fresh ice-cold (0 °C) conical tube and lysates were then precipitated for purification and concentration purposes with 20 % polyethylene glycol (PEG)-8000 (Fisher Scientific, USA), 2.5 M NaCl using a standard protocol and was resuspended in fresh TN buffer. To remove cellular debris, lysates were then filtered through a sterile 0.45 μm syringe filter (BD Discardit, India). To purify lysates from unincorporated fusion and other cellular proteins, particularly unincorporated gpD::eGFP, lysates were purified as previously described by gel chromatography offering lysate purity comparable to that of CsCl centrifugation and amenable to lysate smaller volumes (Zakharova et al. 2005). Briefly, lysates were passed through a 50–150 μL agarose size exclusion column (4 % beads, ABT, Spain) in buffer containing 10 mM Tris–HCl (pH 7.5) and 1 mM MgCl2. Phage were titered at each step of purification by standard viability assays on fresh Sup+ BB4 (supE, supF) E. coli cells, with final phage titers ranging from 1,010 to mid-1,011 phage/ml. Samples were stored at 4 °C.
Sequencing of the λ Dam15 mutation
The λ Dam15 mutation was amplified from λ F7 (λ imm21Dam15) using the primers (F) 5′CACACCAGTGTAAGGGATGTTT -3′; and (R) 5′CCTTTAGTGATGAAGGGTAAAG -3′ (Sigma-Aldrich, Canada). The 330 bp amplified allele was purified and sequenced at York University, North York on an Automated DNA Sequencing Facility on an Applied Biosystems 3130xL DNA Sequencer.
Phage titration and efficiency of plating assays
Viable counts of phage were quantified by standard plaque forming unit assay using BB4 cells (SupE, SupF double suppressor) as the 100 % control as this strain repeatedly generates highest titers of λF7. Plates were incubated overnight at experimental temperature when necessary, otherwise, at 37 °C. Relative plating efficiency at all test temperatures was determined by measuring phage titer on experimental strain divided by that scored on BB4.
Immunoblotting
Immunoblot experiments were conducted using rabbit anti-gfp polyclonal antibody (gift from Dr. B. Moffatt, Waterloo). Samples were denatured by boiling for 10 min, then placed on ice and centrifuged at 12K RPM for 1 min. A total volume of 20 μl was run with a range of 100–200 ng of total protein being run. Samples were run alongside a GFP standard ranging from 10 pg/μl to 100 ng/μl as positive standard, then separated by 15 % SDS-PAGE. After electrophoresis the gel was placed in transfer buffer (48 mM Tris, 39 mM glycine, 20 % (v/v) methanol, 0.04 % (w/v) SDS, (pH 9 to 9.4) for 10 min. The protein was transferred to a nitrocellulose membrane at 20 V for 45 min. The membrane was then stained with Ponceau S stain (0.2 % w/v) by shaking for 10 min and de-staining in dH2O until bands were visible. The stain was then removed by shaking in 1× phosphate-buffered saline (PBS) buffer until the stain (and bands were no longer visible). The membrane was then placed in PVA for 30 s to block, then washed in PBS/milk/Tween for 10 min. The membrane was then incubated at 4 °C overnight in PBS/milk/Tween plus 1/30th dilution of primary rabbit anti-GFP antibody. After incubation the membrane was washed three times in PBS/milk/Tween before adding the secondary AP conjugate antibody (anti-rabbit) at 1:2,000 and incubating for 2 h. The membrane was then rinsed three more times with PBS/milk/Tween for ten min before a final rinse in PBS for five min. The membrane was visualized by a Typhoon imaging system. A second membrane was blocked with TBST for 1 h at room temperature and incubated with primary antibody (1:30) for 1 h at room temperature and washed three times for 10 min with TBST and then incubated with a different secondary antibody in TBST for 1 h at room temperature and then washed again three times and the bands were detected with SuperSignal West Pico Chemiluminescent substrate and visualized on a Kodak imaging system.
Dynamic light scattering of phage
Phage particle size was measured at 25 °C using a Malvern Zetasizer Nano ZS instrument (Malvern instruments, UK). Samples were prepared in Milli-Q water and filtered using a 100 nm filter prior to measurement. The measured sizes are reported using a % intensity distribution. Each data point was automatically repeated in triplicate, and the average is reported. Sizing results are expressed based on “x” increase compared to wild-type, which is λimm21Dam15 (λF7) grown on the BB4 providing gpDwt incorporation into the resultant phage capsid.
Whole phage fluorimetric analysis
Phage samples were prepared in TN buffer by diluting each sample to a uniform concentration of 2.0 × 109 PFU/mL; 150 μL of each prepared sample was then added to a sterile 96 well plate (Starstedt) and were analyzed using a SpectraMax M5 spectrophotometer at an excitation of 485 nm and an emission of 555 nm. The data was analyzed using the SoftMaxPro V5 software where each well was set to be automatically read six times and the samples were run in duplicate with the average being reported. Phage samples were run alongside an eGFP standard (Cell Biolabs Inc. #212103) to determine the protein concentrations of each sample. Phage fluorescence for each preparation derivative was interpolated from the trend-line for known eGFP concentrations. The standard error of each of the sample fluorescence values was done in a weighted analysis against the determination of the sum of the squares of the fluorescence based deviations from the trend-line curve of the fluorescent standard. The standard deviation of each sample was not taken into consideration in the calculations as these deviations were determined to be statistically less significant than the determination from the interpolation itself.
Flow cytometry analysis
Fluorescence and side scatter of phage samples were measured on a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA). Briefly, the flow cytometer was equipped with a 15 mW air-cooled argon-ion laser, with an excitation frequency of 488 nm. Side scatter (SSC) and fluorescence (FL) photomultiplier tube voltages were set to 500 and 525 V, respectively, with logarithmic amplification. A 530/30 nm bandpass filter was used for the observation of D::eGFP fluorescence. All samples were serially diluted 1×, 10×, and 100× using PBS and run for 30 s at the low flow setting (20 μL/min). The dilutions were used to assess instances of “coincidence”, where a high sample concentration results in multiple particles being observed as a single event. More information on how the instrument settings were chosen and the results of negative controls are described by Sokolenko et al. (2012). All data analysis was performed by in-house scripts written in the R programming language (Anon 2011). Samples were compared based on the number of observed fluorescent events (those with FL values greater than 1—the logarithmic scale equivalent of 0) and the distribution of their SSC and FL values. Contour plots of SSC and FL values were generated using kernel density estimation. All plots were created using the ggplot2 package (Wickham 2009).
EDTA sensitivity
Phage were diluted in TN buffer (0.01 M Tris–HCl and 0.1 M NaCl, pH 7.8) (Fisher Scientific, USA) to a universal concentration of 4.0 × 108 PFU/mL then were diluted 100-fold into TN/EDTA (0.01 M Tris–HCl (pH7.8), 0.1 M NaCl, and 0.01 M EDTA) and incubated for 25 min at room temperature. EDTA inactivation was stopped by diluting the samples 100-fold into TN buffer and immediately plating the mixture on BB4 cells (SupE, SupF double suppressor). Plates were incubated overnight at 37 °C. Relative plating efficiency of each sample was determined by measuring phage titer divided by the original phage titers as the 100 % control.
Results
Sequencing the Dam15 mutation and suppressor conferred alleles
Despite the common use of the λF7 (λimm21Dam15) derivatives in phage display, the exact positioning of this mutation has to date never been elucidated. We first sequenced the 333 bp Dam15 allele finding that the amber mutation was localized to the 204th bp of D, converting the 68th CAG codon (glutamine), to an amber translational stop signal (TAG) codon. In a Sup− cell, the amber mutation imparts a premature translational stop resulting in a truncated 68 a.a. non-functional gpD fragment that is incapable of stabilizing and packaging full-length λ DNA. Growing λ Dam15 phage on an amber suppressor host may result in a phenotypic amino acid substitution of glutamine residue at position 68. Only SupE strains will code glutamine at the 68th codon restoring the pristine sequence of gpD, while SupD will confer a serine substitution, yielding gpDQ68S, and SupF will confer a tyrosine substitution, yielding gpDQ68Y. To identify the ability of different amber suppressors to reverse the lethal Dam15 mutation, we generated an isogenic set of amber suppressor derivatives of W3101 (Sup−) and assayed the plating efficiency of λimm21Dam15 (Table 2). Of the three suppressors, the SupD host, yielding gpDQ68S, was least effective at reversing the Dam15 mutation, improving viability by only 10 fold compared to that at Sup− control at 37 °C, and generated pinpoint plaques, indicative of a very low burst size of viable progeny. In contrast, the SupE host that restores the pristine sequence of gpD upon mistranslation of Dam15, restored viability to about 10 % that of the double suppressor (SupE, SupF) positive control. The SupF host, conferring the gpDQ68Y allele performed as well as (if not marginally better than) SupE in an otherwise isogenic host background. SupF restored viability to about 20 % that of the positive control, despite the size and polarity difference between glutamine (SupE) and tyrosine (SupF). These results were corroborated by the efficiency of plating of the strains carrying the parent (backbone) plasmid, pPL451, showing that suppressor capability was not dramatically impacted by differences in temperature, not by the presence of the temperature-regulated expression parent plasmid (Table 2).
Next, we sought to assess the capacity for the pD and pD::eGFP plasmids, in which the expression of the D allele is governed by the temperature-sensitive λ CI857 repressor. The non-suppressor strain W3101 was transformed by pD and pD::eGFP given that the ability to properly package and produce viable λDam15 particles would rely solely upon in trans complementation for the Dam15 mutation from the plasmid. As expected, at increasing temperatures, complementation for Dam15 by plasmid-borne D increased (as repressor activity decreased), with optimal results seen at 39 °C (Table 2) where complementation restored near full viability of λF7. The experimental plasmid expressing D::eGFP also showed a temperature-governed complementation profile that paralleled that of the D plasmid, despite that viability was about tenfold lower at all assayed temperatures, with best complementation achieved again at 39–40 °C. In contrast, the D − parent plasmid was unable to complement for the mutation at any tested temperature.
Complementation for the Dam15 mutation by the gpD and gpD::eGFP plasmids could not be differentiated from suppression of the mutation on SupE and SupF hosts carrying the plasmid due to strong suppressor activity by these strains at all temperatures. In contrast, due to the low viability of λDam15 on the SupD derivative, complementation by gpD::eGFP was observable as expression of the fusion increased with rising temperature and like the Sup− [pD::eGFP] strain, SupD[pD::eGFP] provided the greatest and a similar level of complementation for the Dam15 mutation at 39–40 °C.
Fluorimetric analysis of gpD::eGFP decorated phage
λDam15 phage, variably decorated by gpD::eGFP by passaging through the Sup− and Sup+ strains carrying the pD::eGFP plasmid at various temperatures were standardized for titer and assayed for functional fluorescence by fluorimetry. Fluorescence was interpolated against an eGFP standard of known concentration and the average number of eGFP fusions per phage was determined (Table 3). Due to the lack of expression of a functional gpD allele from either the phage or the plasmid at 30 °C in Sup− [pD::eGFP], a lysate could not be generated under this condition. However, upon raising the temperature to >35 °C, phage viability improved and eGFP decoration was increasingly evident with rising temperature (derepressing expression of D::eGFP) beyond this point, with highest fluorescence observed at 37 °C. All lysate preparations similarly showed increases in fluorescence to 37 °C, albeit considerably lower for phage prepared on the strong suppressor strains, SupE and SupF, which might be attributed to preferential packaging of gpDwt and gpDQ68Y over gpD::eGFP proteins during phage capsid assembly. In contrast, lysates prepared on SupD, the weakest suppressor, demonstrated the strongest fluorescence from all preparations, with a notable signal present even at 30 °C. The noted signal at this temperature was likely due to leaky expression of D::eGFP from the plasmid, but more importantly demonstrates the strong ability of gpD::eGFP to complement for the poorly functional gpDQ68Y protein. We noted quite high variability between readings for SupD prepared lysates at most temperatures, suggesting that despite generating the highest possible incorporation of gpD::eGFP into the phage, decoration between phages prepared on this strain are inconsistent, or may result in multiple preferred decorative species. We also noted that lysates prepared at 39–40 °C indicated decreased fluorescence compared to those prepared at 37 °C on all Sup+ and Sup− strains, despite that derepression of D::eGFP at this temperature is complete and offered highest complementation efficiency for the Dam15 mutation.
Size comparisons of gpD::eGFP decorated phage
Lysates prepared on the Sup series at various temperatures were standardized for titre and sized by dynamic light scattering to approximate relative size differences between undecorated and decorated phage derivatives (Table 4). Sizing of λimm21Dam15 phage grown on SupE, yielding the gpDwt allele, compared well to λimm21 and was used as the wild-type phage size control, generating phage with an average diameter of 62 nm. Relative size differences between λDam15 phage grown on the suppressor host series at 37 °C in the absence of gpD::eGFP, was first determined. As expected, phage grown on Sup− [pD] cells at this inducing temperature were similar in size to the control, while SupD preparations were surprisingly only about a third the size. This finding may suggest that the gpDQ68S protein incorporates very poorly into the capsid, resulting in the smaller capsid size.
Next, λDam15 phage passaged through the suppressors carrying the gpD::eGFP plasmid at various temperatures were sized by DLS in triplicate and compared to the size of the undecorated control phage grown on SupE in the absence of pD::eGFP (Table 4). In all cases, phage size increased as temperature increased from 30 through 37 °C, where expression of D::eGFP was increasingly derepressed. Interestingly, phage samples prepared at 39–40 °C, where D:eGFP expression should be maximal, indicated a 30 to 40 % decrease in phage diameter compared to that at 37 °C, suggesting that either fewer gpD::eGFP were incorporating into the capsid, despite complete derepression of D::eGFP, or eGFP were being shed from the decorated phage. Phage prepared on SupD [pD::eGFP] at 37 and 39 °C were particularly interesting however, as phage samples indicated two notable size peaks, denoting vastly different diameters at substantial occurrence; a first peak at ~30 nm and the second at ~280 nm. The former size is similar to that seen for λF7 passaged through the SupD strain in absence of plasmid. The very large error in average size determination for SupD phage preps at this temperature (Table 4) can be attributed to this bimodal size distribution.
Flow cytometry of gpD::eGFP decorated phage
Lysates prepared on Sup strains at various temperatures were standardized for titre and analyzed with a flow cytometer. A sample’s side scatter and fluorescence profile was found to vary between phage preparations and to be influenced by the degree of eGFP capsid decoration. This group previously optimized the use of flow cytometry for this application using phage passaged through SupE [pD::eGFP] (Sokolenko et al. 2012). A simple count comparison of fluorescent events was the most basic analysis performed on the data obtained (Fig. 1). An overall increase in the count of fluorescent events was observed as a function of temperature for all strains. The temperature dependence was generally more pronounced between 35 and 37 °C than between 30 and 35 °C, especially for SupD and Sup− preparations. For all lysate preparations, the number of fluorescent events evident was an order of magnitude higher between 35 and 37 °C, in agreement with the CI[Ts]857 temperature-lability profile and subsequent expression of gpD::eGFP. Higher culturing temperatures corresponded well with expected higher fluorescence up to 37 °C due to increased gpD::eGFP expression and decoration at this temperature.
Mean fluorescent event counts for phage preparations on Sup−, SupD, SupE and SupF strains carrying pD::eGFP, cultured at 30, 35, 37, and 39 °C. Samples were standardized for titre and diluted tenfold with PBS to avoid coincidence. Fluorescent events are those that had a fluorescence value greater than 1 (the logarithmic scale equivalent of 0). Error bars represent 95 % confidence intervals around the means, calculated from triplicate measurements. In most cases, the error bars are smaller than the size of the corresponding data point
We previously demonstrated that 2D fluorescence/side scatter density is a much better visualization tool for discriminating different types of fluorescent events between preparations. Here, we expanded this approach to compare phage preparations on all Sup strains at all experimental temperatures (Fig. 2). A change in temperature from 30C to 35 °C corresponded to an increase in only side scatter. An increase to 37 °C, on the other hand, had a greater impact on event fluorescence, as can be seen in the prominent movement of the main cluster away from the x-axis. The low temperature changes in side scatter can be interpreted as the result of initial capsid modification, with the increased expression of D::eGFP at 37 °C being required to generate a significant amount of fluorescence. Consistent with the event count analysis, strong increases in Sup− and SupD fluorescence were noted at 37 °C, as compared to the comparatively mild increase in fluorescence observed for lysates prepared on the strong suppressors, SupE and SupF, carrying the gpD::eGFP plasmid. While increasing the culturing temperature to 39 °C did not significantly alter the total number of events detected, it did alter the fluorescence distribution across all preparations, most evident in the decrease of fluorescence for SupD and Sup- strains (Fig. 2). The impact of increasing the expression level of gpD::eGFP by increasing the temperature past 37 °C on the generation of eGFP-tagged phage appears to be limited. Similar to the results obtained for fluorimetry assays, 37 °C again was the upper limit for fluorescence, with evident reductions in fluorescence at 39–40 °C compared to that at 37 °C.
2D density distribution of fluorescence versus side scatter for phage preparations on Sup−, SupD, SupE and SupF strains carrying pD::eGFP, cultured at 30, 35, 37, and 39 °C. The density distribution at each temperature is scaled to a constant height. Contour lines represent fractions of maximum density (density quantiles ranging from 0.10 to 0.80 in intervals of 0.10 and 0.80 to 0.95 in intervals of 0.05). Events with fluorescence or side scatter values of 1 were excluded. Samples were standardized for titre. SupD and Sup− samples cultured at 37 °C had high concentrations that resulted in a considerable amount of coincidence. The contours corresponding to these samples were calculated from samples diluted 10 fold with PBS; all other contours were calculated from undiluted samples to avoid excessive noise
Protein shedding from phage preparations
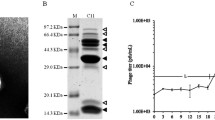
We further investigated the reduction in phage size and fluorescence noted in phages prepared on the Sup series at 40 °C versus their 37 °C counterparts. We first sought to examine whether eGFP were being shed from decorated phage by performing electrophoretic separation, followed by fluorescence analysis on the two species that notes the greatest degree of error for both fluorescence and size determination (Fig. 3). SupD [pD::eGFP] preps demonstrated a high degree of eGFP shedding from the decorated capsid as noted by the strong presence of a ~33 kDa fluorescent protein compared to the ~43 kDa fluorescent protein visualized for gpD::eGFP. This effect was also seen for the Sup− [pD::eGFP] prep. Interestingly, the band runs slightly higher than that seen for eGFP (~29 kDa), which may be due to the incorporation of the linker into the cleaved product. As expected, the SupD [pD] prep phage control, showed no fluorescence as D is not fused to eGFP in this plasmid.
Immunoblot of eGFP species from decorated phage. (A) Phage preparations were standardized for titer and immunoblotted with anti-eGFP. Lane 1 size markers; Lane 2 λimm21Dam15 phage grown on W3101 [pD::eGFP] at 40 °C; Lane 3 λimm21Dam15 phage grown on W3101 supD [pD::eGFP] at 40 °C; Lane 4 λ imm21Dam15 phage grown on W3101 [pD] at 40 °C. (B) eGFP protein standards: 50, 10, 1 ng, respectively
EDTA resistance of decorated and non-decorated phage
To assess the functionality of the surface proteins of λDam15 phage, variably decorated by different gpD::eGFP and/or different isotypes of gpD, we tested each of the phage samples grown at 37 °C for resistance to EDTA (Table 5). We noted a remarkable variation in post-treatment survival rates. Lysates prepared on SupE and on Sup− [pD], both conferring the gpDwt allele, had the greatest EDTA resistance with approximately 91 and 93 % resistant progeny respectively. Phage grown on SupF, generating the gpDQ68Y allele, demonstrated a 44 % survival rate compared to the wild-type, while those grown on SupD, generating the gpDQ68S allele, showed the poorest survival rate of only 35 %. The noted difference in survival rate is believed to be attributed to the relatively stable packaging of gpDwt over either of the gpDQ68Y and gpDQ68S alleles, although gpDQ68Y conferred excellent viability to passaged phage. Phages variably decorated by passaging through Sup+ [pD::eGFP] strains showed relatively consistent survival rates among the samples ranging from 54–59 %, albeit a drop in resistance from that seen with the packaging of gpDwt with SupE and an increase in resistance from those packaging the gpDQ68Y allele from SupF and gpDQ68S alleles from SupD. Lysates developed on Sup− [pD::eGFP] had a relatively low survival rate of 36 % suggesting that decoration is not complete. The variable display of both the fusion protein with pD::eGFP and the gpD alleles conferred were found to be more stable than either of the gpDQ68Y allele, the gpDQ68S allele or the pD::eGFP fusion alone, while those exhibiting wild-type gpD were found to be the most resistant to EDTA, as expected.
Discussion
Maruyama et al. (1994) and Mikawa et al. (1996) first modulated total fusion protein incorporated on the capsid through the use of amber stop codons, located upstream of the gpV/gpD fusion. As such, the level of decoration was dependent on the strength of suppression supplied by the host bacterial Sup+ mutant through which the phage was passaged. These initial studies provided a great foundation and premise for this work while also providing preliminary insight into some of the challenges that would be faced and complications to be overcome. Here, we sought to design and characterize a phage display platform to more finely control decoration of gpD fusions on the surface of λ phage, using eGFP as the fusion partner. This strategy is based on the combination of two competing genetic principles: (1) various alleles of gpD based on isogenic suppression of the λDam15 mutation; and (2) plasmid-borne D::X fusion expression regulated by temperature to complement the Dam15 mutation. We have shown here that through the combination of these two dimensions dramatic variations in phage decoration through gpD::X fusions can be achieved.
The gpD::eGFP C-terminal fusion was able to complement and restore viability to the λDam15 phage, but despite the presence of the linker, the fusion was found to reduce functionality of the major capsid protein marginally in the Sup− host, compared to its unfused counterpart, which is likely due to the size of the fusion generated. Previously, Yang et al. (2000) noted that the presence of a large protein fusion disrupts phage assembly due to the overloading of recombinant protein that decreases phage viability. Vaccaro et al. (2006) later reported that fusion of an scFv to the C-terminal of gpD, employing a similar double gene D system and a flexible linker between the fusion peptide and gpD, similarly resulted in smaller plaque size and reduced phage viability.
In this study, viable phage assembly depended on the tolerance of the phage of the gpD allele conferred from each of the suppressor strains in addition to the gpD::eGFP fusions. While restoration of the pristine sequence (SupE) was expected to restore full function to gpD, it was somewhat surprising that the substitution of the glutamine, a carboxamide, 146.2 Da, polar neutral a.a. with tyrosine, a bulky (181.2 Da), aromatic and hydrophobic a.a. restored nearly full viability to the λDam15 phage. Lastly, the substitution of glutamine with the albeit smaller (105.1 Da) serine that is biochemically similar as a polar neutral a.a. provided very little improvement compared to the Sup− in restoring viability to λ Dam15 and generated comparatively very small phage, and pinpoint plaques in the absence of gpD or gpD::eGFP complementation. Possible reasons for the dramatic effect of the serine substitution at residue 68 of gpD include interference in gpD trimerization and/or compromised interaction with gpE hexamers, although previous findings indicate that the N-terminus is located closer to the threefold axis of the gpD trimer (Yang et al. 2000) and may be involved with interacting with gpE. The functionality of gpD derivatives was similarly reflected in phage sizing of undecorated phage. SupD phage preparations carrying the gpDQ68S allele were only a third that of wild-type and SupF preparations that likely possessed a full complement (405–420 gpD) as compared to SupD phage derivatives that were likely unstable and minimally decorated to restore viability to some progeny, also explaining the poor restoration of phage viability attributed to the gpD68S derivative. To assess stability, we examined EDTA resistance of various preparations in presence and absence of D::eGFP expression and found that phage carrying the gpDQ68S protein were the most unstable. The instability of these phage preparations explains the large variation in eGFP per phage and sizing found for this group, whereby phages are prone to capsid disassembly. This instability was similarly found for phages assembled in the presence of the fusion alone; again indicating that stability requires the presence of gpD to stabilize fusion decoration.
Despite the potential 405 to 420 gpD incorporations into the phage capsid, our results suggest that the highest incorporation possible using our system was only 147 gpD::eGFP molecules/phage. Even in the absence of gpD (Sup−) the incorporation of gpD::eGFP was restricted to 115 molecules per phage, indicating that a full complement of gpD is not necessary to restore viability to λDam15, and that in the absence of a wild-type or gpD derivative, fusion incorporation into the phage is limited. Mikawa et al. (1996) previously reported that a scenario where all gpD sites on the capsid were occupied by C-terminal gpD fusions proteins was not achievable, as resultant phage were still sensitive to EDTA, indicative of empty gpD sites. Moreover, Mikawa et al. (1996) showed that plasmid copy number does affect the level of expression and therefore ability to complement for a D mutation (Mikawa et al. 1996; Sternberg and Hoess 1995). In this work, plasmid copy number was not used as an approach to control decoration and a high copy plasmid was employed for all constructs, but this may be of interest in future studies where multiple fusions could be incorporated simultaneously into a phage capsid. The manipulation of plasmid copy number could provide a third dimension of control. Zucconi et al. (2001) and Minenkova et al. (2003) also found that the number of fusion proteins displayed on the λ capsid surface varied depending on the length and amino acid composition of the foreign sequence expressed at the C-terminal of gpD. Santini et al. (1998) also observed a similar result where fusion of large protein domains to gpD significantly reduced the titre of phage when grown on Sup− compared to Sup+ strains, indicating that incorporating wild-type D protein is preferential for proper assembly, and that the use of suppressor strains is advantageous (Santini et al. 1998). Similarly, Yang et al. (2000) also reported that the interference in phage assembly they noted by overloading the phage with fusion protein was not observed in previous studies where recombinant protein was incorporated with wild-type gpD again suggesting the role of wild-type gpD in improving incorporation of the fusion (Yang et al. 2000).
While suppression correlated well to restoration of phage viability, the opposite was true when we examined fluorescence of the various phage preparations in the presence of D::eGFP expression. Phage prepared on the best suppressors showed low fluorescence even under conditions of high D::eGFP expression, while the SupD suppressor showed the highest levels of eGFP phage decoration, even under conditions where D::eGFP expression was only leaky (30 °C). Previous studies similarly reported that low yields and decreased phage viability are associated with greater expression of capsid-fusion protein, although at the time they could not be sure that this observation was due to impaired capsid assembly (Mikawa et al. 1996; Maruyama et al. 1994).
We attribute this finding to the accumulation of gpD derivatives with higher functionality into the capsid, thereby reducing the incorporation of the less preferred allele. In the case of SupE and SupF preparations, the gpD products are preferred over the gpD::eGFP fusion even at high levels of expression, thereby reducing fluorescence to the resultant phage. In contrast, in SupD preparations, the preferred allele may be gpD::eGFP that “outcompetes” the gpDQ68S allele for placement into the capsid, resulting in stronger decoration. These inferences are supported by phage sizing data, which show a positive correlation between fluorescence and size of the resultant phage particles (Fig. 4). This data supports the assumption that the amount of eGFP fusions on the phage surface can be quantified through florescent measurements and that with an increase in gpD::eGFP fusions, a proportional increase in the phage diameter would be expected. This data was similarly demonstrated by cytometry analysis of fluorescence and side scatter. Here, the 2D fluorescence/side scatter density distribution (Fig. 2) corroborated fusion decoration trends and as previously suggested (Sokolenko et al. 2012) indicated increasing side scatter alongside heightened fluorescence (eGFP decoration), relating surface complexity to degree of fusion decoration.
Separation of decorated phage preparations based on fluorescence emitted and size as determined by DLS. Fluorescence and size ratios based on comparison to λF7 grown on W3101 supE [pD::eGFP]. The measured sizes are reported using a % intensity distribution. Each data point was automatically repeated in triplicate, and the average is reported. Sizing results are expressed based on “x” increase compared to wild-type, which is λimm21Dam15 (λF7) grown on the BB4 providing gpDwt incorporation into the resultant phage capsid. Measured fluorescence data was analyzed using the SoftMaxPro V5 software based on six readings and the samples were run in duplicate with the average being reported. Phage samples were run alongside an eGFP standard to determine the protein concentrations of each sample. Phage fluorescence for each preparation derivative was interpolated from the trend-line for known eGFP concentrations
While the eGFP phage decoration profile fits well for increasing D::eGFP derepression from 30° through 37 °C, at the highest level of D::eGFP expression from λ pL (39–40 °C), where CI857 repressor is completely labile, eGFP phage decoration decreases compared to that at 37 °C for all cell line preparations. In addition, phage prepared on hosts under this state of metabolic stress indicated a high degree of breakage, or cleavage of eGFP from the phage head (Fig. 3). We attribute this finding to the mode of lysate preparation that exposes host cells to heat shock temperatures for several hours, altering cell physiology and increasing concentration of heat shock response proteins (hsp), controlled by σ32 factor (Smith 2007; Gill et al. 2000; Valdez-Cruz et al. 2010). These include the upregulation of genes coding for heat shock proteins and SOS response, with the particular attention to proteases such as clpP, degP, ftsH, ompT, and lon. Temperature inducing up-shifts impart high physiological stress to the host cell that results in a threefold increase in global protein synthesis (a quarter of which is devoted to hsp synthesis) (Wegrzyn and Wegrzyn 2002; Hoffmann and Rinas 2001; Zhao et al. 2005; Yamamori and Yura 1980; Hoffmann and Rinas 2000; Valdez-Cruz et al. 2010), thereby reducing growth rates and recombinant protein yield (Wegrzyn and Wegrzyn 2002; Valdez-Cruz et al. 2010; Zhao et al. 2005). Lastly, recombinant proteins produced using thermoinducing systems, particularly from multicopy plasmids as in this work, can result in high protein concentrations and the formation of inclusion bodies (Babu et al. 2000; Rinas and Hoffmann 2004; Caspeta et al. 2009; Vallejo et al. 2002; Valdez-Cruz et al. 2010).
In an effort to tweak our system, we examined and report elsewhere, the influence of various hsp and SOS proteases on eGFP phage decoration at 40 °C, and found that an otherwise isogenic lon mutant host conferred a dramatically improved yield under heat stress conditions. Similarly, limited induction shifts of 1 h from 37 °C to 42 °C dramatically improved decoration at 40 °C as previously demonstrated (Aucoin et al. 2006).
The potential downstream applications of this research are the construction of multivalent phage vaccines and/or the development of vehicles for phage-mediated targeted gene therapy. These applications exhibit the two endpoints of the spectrum of phage surface display control. While vaccination would expectedly be most effective if display saturation was reached, targeted transgene delivery applications would require a high-level of specificity in targeting unique receptors of appropriate cells, where fewer and controlled fusions are usually preferable. It is important to note that while our system demonstrates a fair degree of variability in eGFP decoration, these results are only useful in relative comparison of phage decoration conditions. Absolute display is dependent upon the size and biochemical attributes of the fused candidate and the tolerance of gpD and the foreign polypeptide to fusion.
References
Anon, 2011. R Development Core Team. R: A language and environment for statistical computing. R: A language and environment for statistical computing. Available at: http://www.r-project.org. Accessed 7 January 2013.
Aucoin MG, McMurray-Beaulieu V, Poulin F, Boivin EB, Chen J, Ardelean FM, Cloutier M, Choi YJ, Miguez CB, Jolicoeur M (2006) Identifying conditions for inducible protein production in E. coli: combining a fed-batch and multiple induction approach. Microbial cell factories 5(27)
Babu KR, Swaminathan S, Marten S, Khanna N, Rinas U (2000) Production of interferon-a in high cell density cultures of recombinant Escherichia coli and its single step puri ® cation from refolded inclusion body proteins. Appl Microbiol Biotechnol 53:655–660. doi:10.1007/s002530000318
Bachmann BJ (1972) Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev 36(4):525–557
Beghetto E, Pucci A, Minenkova O, Spadoni A, Bruno L, Buffolano W, Soldati D, Felici F, Gargano N (2001) Identification of a human immunodominant B-cell epitope within the GRA1 antigen of Toxoplasma gondii by phage display of cDNA libraries. Internal J Parasitol 31(14):1659–1668. doi:10.1016/S0020-7519(01)00288-0
Beghetto E, Gargano N (2011) Lambda-display: a powerful tool for antigen discovery. Mol 16(4):3089–3105. doi:10.3390/molecules16043089
Caspeta L, Noemí F, Néstor OP, Francisco B, Octavio TR (2009) The effect of heating rate on Escherichia coli metabolism, physiological stress, transcriptional response, and production of temperature-induced recombinant protein: a scale-down study. Biotechnol Bioeng 102(2):468–482. doi:10.1002/bit.22084
Dokland T, Murialdo H (1993) Structural transitions during maturation of Lambda. J Mol Biol 233:682–694. doi:10.1006/jmbi.1993.1545
Dunn IS (1995) Assembly of functional bacteriophage lambda virions incorporating C-terminal peptide or protein fusions with the major tail protein. J Mol Biol 248(3):497–506. doi:10.1006/jmbi.1995.0237
Edlin JD, Sundaram TK (1989) Regulation of isocitrate dehydrogenase by phosphorylation in Escherichia coli K-12 and a simple method for determining the amount of inactive phosphoenzyme. J Bacteriol 171(5):2634–2638
Georgopoulos C, Tilly K, Casjens S (1983) Lambdoid Phage Head Assembly. In: Hendrix R et al (eds) Lambda II edn. Cold Springs Harbor, New York, pp 279–304
Gill RT, Valdes JJ, Bentley WE (2000) A comparative study of global stress gene regulation in response to overexpression of recombinant proteins in Escherichia coli. Metab Eng 2(3):178–189. doi:10.1006mben.2000.0148
Grossman AD, Erickson JW, Gross CA, Singer M, Baker TA, Schnitzler G, Deischel SM et al (1989) A collection of strains containing genetically for genetic mapping of Escherichia coli. Microbiol Mol Biol R 53(1):1–24
Gupta A, Onda M, Pastan I, Adhya S, Chaudhary VK (2003) High-density functional display of proteins on bacteriophage lambda. J Mol Biol 334(2):241–254. doi:10.1016/j.jmb.2003.09.033
Hayes S, Gamage LN, Hayes C (2010) Dual expression system for assembling phage lambda display particle (LDP) vaccine to porcine Circovirus 2 (PCV2). Vaccine 28(41):6789–6799. doi:10.1016/j.vaccine.2010.07.047
Hoffman EP, Wilhelm RC (1970) Genetic mapping and dominance of the amber suppressor, Su1 (supD), in Escherichia coli K-12. J Bacteriol 103(1):32–36
Hoffmann F, Rinas U (2000) Kinetics of heat-shock response and inclusion body formation during temperature-induced production of basic fibroblast growth factor in high-cell-density cultures of recombinant Escherichia coli. Biotechnol Prog 16(6):1000–1007. doi:10.1021/bp0000959
Hoffmann F, Rinas U (2001) On-Line estimation of the metabolic burden resulting from the synthesis of plasmid-encoded and heat-shock proteins by monitoring respiratory energy generation. Biotechnol Bioeng 76(4):333–340. doi:10.1002/bit.10098
Kasman LM, Lukowiak AA, Garczynski SF, McNall RJ, Youngman P, Adang MJ (1998) Phage display of a biologically active Bacillus thuringiensis toxin. Appl Envi Micro 64(8):2995–3003
Kehoe JW, Kay BK (2005) Filamentous Phage display in the new millennium. Chem Rev 3824:4056–4072. doi:10.1021/cr000261r
Larocca D, Burg MA, Jensen-Pergakes K, Ravey EP, Gonzalez AM, Baird A (2002) Evolving phage vectors for cell targetted gene delivery. Current Pharmaceut Biotechnol 3(1):45–57. doi:10.2174/1389201023378490
Maruyama IN, Maruyama HI, Brenner S (1994) Lambda foo: a lambda phage vector for the expression of foreign proteins. Proc National Acad Sci USA 91(17):8273–8277
Marzari R, Edomi P, Bhatnagar RK, Ahmad S, Selvapandiyan A, Bradbury A (1997) Phage display of Bacillus thuringiensis CrylA (A) insecticidal toxin. FEBS Lett 411(1):27–31
Mikawa YG, Maruyama IN, Brenner S (1996) Surface display of proteins on bacteriophage lambda heads. J Mol Biol 262(1):21–30. doi:10.1006/jmbi.1996.0495
Minenkova O, Pucci A, Pavoni E, De Tomassi A, Fortugno P, Gargano N, Cianfriglia M et al (2003) Identification of tumor-associated antigens by screening phage-displayed human cDNA libraries with sera from tumor patients. Int J Cancer 106(4):534–544. doi:10.1002/ijc.11269
Nichols BP, Shafiq O, Meiners V (1998) Sequence analysis of Tn 10 insertion sites in a collection of Escherichia coli strains used for genetic mapping and strain construction sequence. J Bacteriol 180(23):6408–6411
Nilsson F, Tarli L, Viti F, Neri D (2000) The use of phage display for the development of tumour targeting agents. Adv Drug Deliv Rev 43(2–3):165–196. doi:10.1016/S0169-409X(00)00068-5
Petty NK, Evans TJ, Fineran PC, Salmond GPC (2007) Biotechnological exploitation of bacteriophage research. Trends Biotechnol 25(1):7–15. doi:10.1016/j.tibtech.2006.11.003
Ptashne M (2004) A genetic switch—phage lambda revisited 3rd edn. Cold Springs Harbor, New York
Rinas U, Hoffmann F (2004) Selective leakage of host-cell proteins during high-cell-density cultivation of recombinant and non-recombinant Escherichia coli. Biotechnol Prog 20(3):679–687. doi:10.1021/bp034348k
Santini C, Brennan D, Mennuni C, Hoess RH, Nicosia A, Cortese R, Luzzago A (1998) Efficient display of an HCV cDNA expression library as C-terminal fusion to the capsid protein D of bacteriophage lambda. J Mol Biol 282(1):125–135. doi:10.1006/jmbi.1998.1986
Seow Y, Wood MJ (2009) Biological gene delivery vehicles: beyond viral vectors. Mol Ther 17(5):767–777. doi:10.1038/mt.2009.41
Smith GP (1985) Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion. Adv Sci 228(4705):1315–1317
Smith HE (2007) The transcriptional response of Escherichia coli to recombinant protein insolubility. J Struct Funct Genomics 8(1):27–35. doi:10.1007/s10969-007-9030-7
Sokolenko S, Nicastro J, Slavcev R, Aucoin MG (2012) Graphical analysis of flow cytometer data for characterizing controlled fluorescent protein display on λ phage. Cytom Part A : J Int Soc Anal Cyt 81(12):1–9. doi:10.1002/cyto.a.22211
Sternberg N, Hoess RH (1995) Display of peptides and proteins on the surface of bacteriophage lambda. Proc National Acad Sci USA 92(5):1609–1613. doi:10.1073/pnas.92.5.1609
Sternberg N, Hamilton D, Enquist L, Weisberg R (1979) A simple technique for the isolation of deletion mutants of phage lambda. Cancer Res 8:35–51
Vaccaro P, Pavoni E, Monteriù G, Andrea P, Felici F, Minenkova O (2006) Efficient display of scFv antibodies on bacteriophage lambda. J Immunol Methods 310(1–2):149–158. doi:10.1016/j.jim.2006.01.003
Valdez-Cruz NA, Caspeta L, Pérez NO, Ramírez OT, Trujillo-Roldán MA (2010) Production of recombinant proteins in E. coli by the heat inducible expression system based on the phage lambda pL and/or pR promoters. Microbial Cell fact 9(18). doi: 10.1186/1475-2859-9-18
Vallejo LF, Brokelmann M, Marten S, Trappe S, Cabrera-Crespo J, Hoffmann A, Gross G, Weich HA, Rinas U (2002) Renaturation and purification of bone morphogenetic protein-2 produced as inclusion bodies in high-cell-density cultures of recombinant Escherichia coli. J Biotechnol 94(2):185–194. doi:10.1016/S0168-1656(01)00425-4
Vilchez S, Jacoby J (2004) Display of biologically functional insecticidal toxin on the surface of lambda phage. Appl Environ Microbiol 70(11):6587–6594. doi:10.1128/AEM.70.11.6587-6594.2004
Wegrzyn G, Wegrzyn A (2002) Stress responses and replication of plasmids in bacterial cells. Microbial Cell fact 1(1):2. doi:10.1186/1475-2859-1-2
Wickham H (2009) ggplot2: elegant graphics for data analysis; 2nd printing. Springer, New York
Willats WGT (2002) Phage display: practicalities and prospects. Plant Mol Biol 50:837–854
Windass JD, Brammar WJ (1979) Aberrant immunity behaviour of hybrid of ColE1-type plasmids phages containing the DNA. MGG 172(3):329–337
Yamamori T, Yura T (1980) Temperature-induced synthesis of specific proteins in Escherichia coli: evidence for transcriptional control. J Bacteriol 142(3):843–851
Yang F, Forrer P, Dauter Z, Conway JF, Cheng N, Cerritelli ME, Steven AC, Plückthun A, Wlodawer A (2000) Novel fold and capsid-binding properties of the λ-phage display platform protein gpD. Nat Struct Biol 7(3):230–237
Zakharova MY, Kozyr AV, Ignatova AN, Vinnikov IA, Shemyakin IG, Kolesnikov AV (2005) Purification of filamentous bacteriophage for phage display using size-exclusion chromatography. BioTechniques 38(2):194, 196, 198
Zanghi CN, Lankes HA, Bradel-Tretheway B, Wegman J, Dewhurst S (2005) A simple method for displaying recalcitrant proteins on the surface of bacteriophage lambda. Nucleic acids Res 33(18):e160. doi: 10.1093/nar/gni158 http://nar.oxfordjournals.org/content/33/18/e160.short - aff-1
Zanghi CN, Sapinoro R, Bradel-Tretheway B, Dewhurst S (2007) A tractable method for simultaneous modifications to the head and tail of bacteriophage lambda and its application to enhancing phage-mediated gene delivery. Nucleic Acids Res 35(8):e59. doi:10.1093/nar/gkm146
Zhao K, Liu M, Burgess RR (2005) The global transcriptional response of Escherichia coli to induced sigma 32 protein involves sigma 32 regulon activation followed by inactivation and degradation of sigma 32 in vivo. J Biol Chem 280(18):17758–17768. doi:10.1074/jbc.M500393200
Zucconi A, Dente L, Santonico E, Castagnoli L, Cesareni G (2001) Selection of ligands by panning of domain libraries displayed on phage lambda reveals new potential partners of synaptojanin 1. J Mol Biol 307(5):1329–1339. doi:10.1006/jmbi.2001.4572
Acknowledgments
This work was supported by UW Start-up funds to RAS; and Drug Safety and Effectiveness Cross-Disciplinary Training (DSECT) Scholarship provided by Canadian Institute of Health Research (CIHR) to JN.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Nicastro, J., Sheldon, K., El-zarkout, F.A. et al. Construction and analysis of a genetically tuneable lytic phage display system. Appl Microbiol Biotechnol 97, 7791–7804 (2013). https://doi.org/10.1007/s00253-013-4898-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-013-4898-6