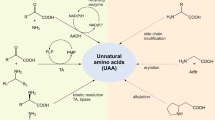
Abstract
The great importance of amide bonds in industrial synthesis has encouraged the search for efficient catalysts of amide bond formation. Microbial transglutaminase (MTG) is heavily utilized in crosslinking proteins in the food and textile industries, where the side chain of a glutamine reacts with the side chain of a lysine, forming a secondary amide bond. Long alkylamines carrying diverse chemical entities can substitute for lysine as acyl-acceptor substrates, to link molecules of interest onto peptides or proteins. Here, we explore short and chemically varied acyl-acceptor substrates, to better understand the nature of nonnatural substrates that are tolerated by MTG, with the aim of diversifying biocatalytic applications of MTG. We show, for the first time, that very short-chain alkyl-based amino acids such as glycine can serve as acceptor substrates. The esterified α-amino acids Thr, Ser, Cys, and Trp—but not Ile—also showed reactivity. Extending the search to nonnatural compounds, a ring near the amine group—particularly if aromatic—was beneficial for reactivity, although ring substituents reduced reactivity. Overall, amines attached to a less hindered carbon increased reactivity. Importantly, very small amines carrying either the electron-rich azide or the alkyne groups required for click chemistry were highly reactive as acyl-acceptor substrates, providing a robust route to minimally modified, “clickable” peptides. These results demonstrate that MTG is tolerant to a variety of chemically varied natural and nonnatural acyl-acceptor substrates, which broadens the scope for modification of Gln-containing peptides and proteins.


Similar content being viewed by others
References
Abe H, Goto M, Kamiya N (2010) Enzymatic single-step preparation of multifunctional proteins. Chem Commun 46:7160–7162
Best MD (2009) Click chemistry and bioorthogonal reactions: unprecedented selectivity in the labeling of biological molecules. Biochemistry 48:6571–6584
Clouthier CM, Pelletier JN (2012) Expanding the organic toolbox: a guide to integrating biocatalysis in synthesis. Chem Soc Rev 41:1585–1605
Fichert T, Yazdanian M, Proudfoot JR (2003) A structure–permeability study of small drug-like molecules. Bioorg Med Chem Lett 13:719–722
Fleming FF, Yao L, Ravikumar PC, Funk L, Shook BC (2010) Nitrile-containing pharmaceuticals: efficacious roles of the nitrile pharmacophore. J Med Chem 53:7902–7917
Folk JE, Cole PW (1966) Transglutaminase: mechanistic features of the active site as determined by kinetic and inhibitor studies. Biochim Biophys Acta Enzymol Biol Oxid 122:244–264
Gnaccarini C, Ben-Tahar W, Mulani A, Roy I, Lubell WD, Pelletier JN, Keillor JW (2012) Site-specific protein propargylation using tissue transglutaminase. Org Biomol Chem 10:5258–5265
Hernandes MZ, Cavalcanti SMT, Moreira DRM, De Azevedo Junior WF, Leite ACL (2010) Halogen atoms in the modern medicinal chemistry: hints for the drug design. Current Drug Targets 11:303–314
Hilal SH, Karickhoff SW, Carreira LA (1995) A rigorous test for SPARC’s chemical reactivity models: estimation of more than 4300 ionization pKa’s. Quant Struc Act Rel 14:348–355
Himo F, Lovell T, Hilgraf R, Rostovtsev VV, Noodleman L, Sharpless KB, Fokin VV (2005) Copper(I)-catalyzed synthesis of azoles. DFT study predicts unprecedented reactivity and intermediates. J Am Chem Soc 127:210–216
Inverarity IA, Hulme AN (2007) Marked small molecule libraries: a truncated approach to molecular probe design. Org Biomol Chem 5:636–643
Jeger S, Zimmermann K, Blanc A, Grunberg J, Honer M, Hunziker P, Struthers H, Schibli R (2010) Site specific and stoichiometric modification of antibodies by bacterial transglutaminase. Angew Chem Int Ed 49:1–4
Kashiwagi T, Yokoyama K-I, Ishikawa K, Ono K, Ejima D, Matsui H, Suzuki E (2002) Crystal structure of microbial transglutaminase from Streptoverticillium mobaraense. J Biol Chem 277:44252–44260
Keillor JW, Chica RA, Chabot N, Vinci V, Pardin C, Fortin E, Gillet SMFG, Nakano Y, Kaartinen MT, Pelletier JN, Lubell WD (2008) The bioorganic chemistry of transglutaminase—from mechanism to inhibition and engineering. Can J Chem 86:271–276
Kitaoka M, Tsuruda Y, Tanaka Y, Goto M, Mitsumori M, Hayashi K, Hiraishi Y, Miyawaki K, Noji S, Kamiya N (2011) Transglutaminase-mediated synthesis of a DNA-(enzyme)n probe for highly sensitive DNA detection. Chemistry 17:5387–5392
Kolb HC, Finn MG, Sharpless KB (2001) Click chemistry: diverse chemical function from a few good reactions. Angew Chem Int Ed 40:2004–2021
Le GT, Abbenante G, Madala PK, Hoang HN, Fairlie DP (2006) Organic azide inhibitors of cysteine proteases. J Am Chem Soc 128:12396–12397
Leblanc A, Gravel C, Labelle J, Keillor JW (2001) Kinetic studies of guinea pig liver transglutaminase reveal a general-base-catalyzed deacylation mechanism. Biochemistry 40:8335–8342
Lee J-H, Song C, Kim D-H, Park I-H, Lee S-G, Lee Y-S, Kim B-G (2013) Glutamine (Q)-peptide screening for transglutaminase reaction using mRNA display. Biotechnol Bioeng 110:353–362
Marx CK, Hertel TC, Pietzsch M (2007) Soluble expression of a pro-transglutaminase from Streptomyces mobaraensis in Escherichia coli. Enzyme Microb Technol 40:1543–1550
Mero A, Schiavon M, Veronese FM, Pasut G (2011) A new method to increase selectivity of transglutaminase mediated PEGylation of salmon calcitonin and human growth hormone. J Controlled Release 154:27–34
Mindt TL, Jungi V, Wyss S, Friedli A, Pla G, Novak-Hofer I, Grünberg J, Schibli R (2008) Modification of different IgG1 antibodies via glutamine and lysine using bacterial and human tissue transglutaminase. Bioconjugate Chem 19:271–278
Ohtsuka T, Ota M, Nio N, Motoki M (2000a) Comparison of substrate specificities of transglutaminases using synthetic peptides as acyl donors. Biosci Biotechnol Biochem 64:2608–2613
Ohtsuka T, Sawa A, Kawabata R, Nio N, Motoki M (2000b) Substrate specificities of microbial transglutaminase for primary amines. J Agric Food Chem 48:6230–6233
Park KD, Morieux P, Salomé C, Cotten SW, Reamtong O, Eyers C, Gaskell SJ, Stables JP, Liu R, Kohn H (2009) Lacosamide isothiocyanate-based agents: novel agents to target and identify lacosamide receptors. J Med Chem 52:6897–6911
Pasternack R, Dorsch S, Otterbach JT, Robenek IR, Wolf S, Fuchsbauer HL (1998) Bacterial pro-transglutaminase from Streptoverticillium mobaraense—purification, characterisation and sequence of the zymogen. Eur J Biochem/FEBS 257:570–576
Pasternack R, Laurent HP, Rüth T, Kaiser A, Schön N, Fuchsbauer HL (1997) A fluorescent substrate of transglutaminase for detection and characterization of glutamine acceptor compounds. Anal Biochem 249:54–60
Pattabiraman VR, Bode JW (2011) Rethinking amide bond synthesis. Nature 480:471–479
Porta R, Di Pierro P, Sorrentino A, Mariniello L (2011) Promising perspectives for transglutaminase in “bioplastics” production. J Biotechnol Biomaterial 01:1–4
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor
Schmid A, Dordick JS, Hauer B, Kiener A, Wubbolts M, Witholt B (2001) Industrial biocatalysis today and tomorrow. Nature 409:258–268
Studier FW (2005) Protein production by auto-induction in high-density shaking cultures. Protein Expression Purif 41:207–234
Sugimura Y, Yokoyama K, Nio N, Maki M, Hitomi K (2008) Identification of preferred substrate sequences of microbial transglutaminase from Streptomyces mobaraensis using a phage-displayed peptide library. Arch Biochem Biophys 477:379–383
Tagami U, Shimba N, Nakamura M, Yokoyama K-I, Suzuki E-I, Hirokawa T (2009) Substrate specificity of microbial transglutaminase as revealed by three-dimensional docking simulation and mutagenesis. Protein Eng Des Sel 22:747–752
Tanaka T, Kamiya N, Nagamune T (2004) Peptidyl linkers for protein heterodimerization catalyzed by microbial transglutaminase. Bioconjugate Chem 15:491–497
Tanaka T, Kamiya N, Nagamune T (2005) N-terminal glycine-specific protein conjugation catalyzed by microbial transglutaminase. FEBS Lett 579:2092–2096
Tominaga J, Kemori Y, Tanaka Y, Maruyama T, Kamiya N, Goto M (2007) An enzymatic method for site-specific labeling of recombinant proteins with oligonucleotides. Chem Commun 401–3
Umezawa Y, Ohtsuka T, Yokoyama K (2002) Comparison of enzymatic properties of microbial transglutaminase from Streptomyces sp. Food Sci Technol Res 8:113–118
Valeur E, Bradley M (2009) Amide bond formation: beyond the myth of coupling reagents. Chem Soc Rev 38:606–631
Yokoyama K, Nio N, Kikuchi Y (2004) Properties and applications of microbial transglutaminase. Appl Microbiol Biotechnol 64:447–454
Zhu Y, Tramper J (2008) Novel applications for microbial transglutaminase beyond food processing. Trends Biotechnol 26:559–565
Acknowledgments
This work was supported by funding from the Natural Sciences and Engineering Research Council of Canada and CGCC, the Center in Green Chemistry and Catalysis. We thank Fabrice Galaud and Pierre Lavallée, in the Combinatorial Chemistry Laboratory at the Université de Montréal, for their generous technical help, Alexandra Furtos in the Regional Mass Spectrometry Center at Université de Montréal for her assistance, and Maria Stoïca and Dr. Christophe Pardin for the synthesis of azide compounds and acknowledge Klavdja Annabel Fignolé for assisting in experimental work and Natalie Rachel for commenting and revising the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 36 kb)
Rights and permissions
About this article
Cite this article
Gundersen, M.T., Keillor, J.W. & Pelletier, J.N. Microbial transglutaminase displays broad acyl-acceptor substrate specificity. Appl Microbiol Biotechnol 98, 219–230 (2014). https://doi.org/10.1007/s00253-013-4886-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-013-4886-x