Abstract
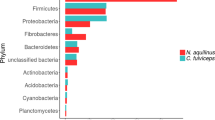
Decomposition of lignocelluloses by cooperative microbial actions is an essential process of carbon cycling in nature and provides a basis for biomass conversion to fuels and chemicals in biorefineries. In this study, structurally stable symbiotic aero-tolerant lignocellulose-degrading microbial consortia were obtained from biodiversified microflora present in industrial sugarcane bagasse pile (BGC-1), cow rumen fluid (CRC-1), and pulp mill activated sludge (ASC-1) by successive subcultivation on rice straw under facultative anoxic conditions. Tagged 16S rRNA gene pyrosequencing revealed that all isolated consortia originated from highly diverse environmental microflora shared similar composite phylum profiles comprising mainly Firmicutes, reflecting convergent adaptation of microcosm structures, however, with substantial differences at refined genus level. BGC-1 comprising cellulolytic Clostridium and Acetanaerobacterium in stable coexistence with ligninolytic Ureibacillus showed the highest capability on degradation of agricultural residues and industrial pulp waste with CMCase, xylanase, and β-glucanase activities in the supernatant. Shotgun pyrosequencing of the BGC-1 metagenome indicated a markedly high relative abundance of genes encoding for glycosyl hydrolases, particularly for lignocellulytic enzymes in 26 families. The enzyme system comprised a unique composition of main-chain degrading and side-chain processing hydrolases, dominated by GH2, 3, 5, 9, 10, and 43, reflecting adaptation of enzyme profiles to the specific substrate. Gene mapping showed metabolic potential of BGC-1 for conversion of biomass sugars to various fermentation products of industrial importance. The symbiotic consortium is a promising simplified model for study of multispecies mechanisms on consolidated bioprocessing and a platform for discovering efficient synergistic enzyme systems for biotechnological application.






Similar content being viewed by others
References
Allgaier M, Reddy A, Park JI, Ivanova N, D’haeseleer P, Lowry S, Sapra R, Hazen TC, Simmons BA, Vander Gheynst JS, Hugenholtz P (2010) Targeted discovery of glycoside hydrolases from a switchgrass-adapted compost community. PLoS One 5:e8812
Alvira P, Negro MJ, Ballesteros M (2011) Effect of endoxylanase and α-L-arabinofuranosidase supplementation on the enzymatic hydrolysis of steam exploded wheat straw. Bioresour Technol 102:4552–4558
Brulc JM, Antonopoulos DA, Miller MEB, Wilson MK, Yannarell AC, Dinsdale EA, Edwards RE, Frankh ED, Emersoni JB, Wacklini P, Coutinhoj PM, Henrissatj B, Nelsoni KE, White BA (2009) Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc Nat Acad Sci USA 106:1946–1953
Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233–238
Chen S, Dong X (2004) Acetanaerobacterium elongatum gen. nov., sp. nov., from paper mill waste water. Inter J Sys Evol Microbiol 54:2257–2262
Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Crespo CF, Badshah M, Alvarez MT, Mattiasson B (2012) Ethanol production by continuous fermentation of D-(+)-cellobiose, D-(+)-xylose and sugarcane bagasse hydrolysate using the thermoanaerobe Caloramator boliviensis. Bioresour Technol 103:186–191
Doi RH (2008) Cellulases of mesophilic microorganisms cellulosome and noncellulosome producers. Ann NY Acad Sci 1125:267–279
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
FitzPatrick M, Champagne P, Cunningham MF, Whitney RA (2010) A biorefinery processing perspective: treatment of lignocellulosic materials for the production of value-added products. Bioresour Technol 101:8915–8922
Fengel D, Wegener G (1984) Wood: chemistry, ultrastructure, reactions. De Gruyter, Berlin, Germany
Feng Y, Yu Y, Wang X, Qu Y, Li D, He W, Kim BH (2011) Degradation of raw corn stover powder (RCSP) by an enriched microbial consortium and its community structure. Bioresour Technol 102:742–747
Fu Z, Holtzapple MT (2010) Consolidated bioprocessing of sugarcane bagasse and chicken manure to ammonium carboxylates by a mixed culture of marine microorganisms. Bioresour Technol 101:2825–2836
Guo P, Zhu W, Wang H, Lü Y, Wang X, Zheng D, Cui Z (2010) Functional characteristics and diversity of a novel lignocelluloses degrading composite microbial system with high xylanase activity. J Microbiol Biotechnol 20:254–264
Haruta S, Cui Z, Huang Z, Li M, Ishii M, Igarashi Y (2002) Construction of a stable microbial community with high cellulose-degradation ability. Appl Microbiol Biotechnol 59:529–534
He Q, Hemme CL, Jiang H, He Z, Zhou J (2011) Mechanisms of enhanced cellulosic bioethanol fermentation by co-cultivation of Clostridium and Thermoanaerobacter spp. Bioresour Technol 102:9586–9592
Kanokratana P, Uengwetwanit T, Rattanachomsri U, Bunterngsook B, Nimchua T, Tangphatsornruang S, Plengvidhaya V, Champreda V, Eurwilaichitr L (2010) Metagenomic analysis on phylogenetics and metabolic potential of microbial community in primary tropical peat swamp forest. Microb Ecol 61:518–528
Kato S, Haruta S, Cui ZJ, Ishii M, Igarashi Y (2004) Effective cellulose degradation by a mixed-culture system composed of a cellulolytic Clostridium and aerobic non-cellulolytic bacteria. FEMS Microbiol Ecol 51:133–142
Kato S, Haruta S, Cui ZJ, Ishii M, Igarashi Y (2005) Stable coexistence of five bacterial strains as a cellulose-degrading community. Appl Environ Microbiol 71:7099–7106
Kato S, Haruta S, Cui ZJ, Ishii M, Igarashi Y (2008) Network relationships of bacteria in a stable mixed culture. Microb Ecol 56:403–411
Li L-L, McCorkle SR, Monchy S, Taghavi S, van der Lelie D (2009) Bioprospecting metagenomes: glycosyl hydrolases for converting biomass. Biotechnol Biofuel 2:10
Lu Y, Zhang Y, Lynd LR (2006) Enzyme-microbe synergy during cellulose hydrolysis by Clostridium thermocellum. Proc Nat Acad Sci USA 103:16165–16169
Lukashin AV, Borodovsky M (1998) GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res 26:1107–1115
Lynd LR, Weimer PJ, Van Zyl WH, Pretorius IS (2002) Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev 66:506–739
Meyer M, Stenzel U, Hofreiter M (2008) Parallel tagged sequencing on the 454 platform. Nat Protoc 3:267–278
Mitchell WJ (1998) Physiology of carbohydrate to solvent conversion by clostridia. Adv Microb Physiol 39:31–130
Narisawa N, Haruta S, Cui ZJ, Ishii M, Igarashi Y (2007) Effect of adding cellulolytic bacterium on stable cellulose-degrading microbial community. J Biosci Bioeng 104:432–434
Nimchua T, Thongaram T, Uengwetwanit T, Pongpattanakitshote S, Eurwilaichitr L (2011) Metagenomic analysis of novel lignocellulose-degrading enzymes from higher termite guts inhabiting microbes. J Microbiol Biotechnol 22:462–469
Okura N, Soneura M, Ninomiya K, Katakura Y, Shioya S (2008) Biological detoxification of waste house wood hydrolysate using Ureibacillus thermosphaericus for bioethanol production. J Biosci Bioeng 106:128–133
Pope PB, Denman SE, Jones M, Tringe SG, Barry K, Malfatti SA, McHardy AC, Cheng JF, Hugenholtz P, McSweeney CS, Morrison M (2010) Adaptation of herbivory by the Tammar wallaby includes bacterial and glycoside hydrolase profiles different from other herbivores. Proc Nat Acad Sci 107:14793–14798
Rattanachomsri U, Tanapongpipat S, Eurwilaichitr L, Champreda V (2009) Simultaneous non-thermal saccharification of cassava pulp by multi-enzyme activity and ethanol fermentation by Candida tropicalis. J Biosci Bioeng 107:488–493
Ren N, Xing D, Rittmann BE, Zhao L, Xie T, Zhao X (2007) Microbial community structure of ethanol type fermentation in bio-hydrogen production. Environ Microbiol 9:1112–1125
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
TAPPI (1992) TAPPI test method: determination of cellulose and pentosan contents of wood. Technical Association of Pulp and Paper Industry (TAPPI) Press, Atlanta
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Wang W, Yan L, Cui Z, Gao Y, Wang Y, Jing R (2011) Characterization of a microbial consortium capable of degrading lignocellulose. Bioresour Technol 102:9321–9324
Warnecke F, Luginbühl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT, Cayouette M, McHardy AC, Djordjevic G, Aboushadi N, Sorek R, Tringe SG, Podar M, Martin HG, Kunin V, Dalevi D, Madejska J, Kirton E, Platt D, Szeto E, Salamov A, Barry K, Mikhailova N, Kyrpides NC, Matson EG, Ottesen EA, Zhang X, Hernandez M, Murillo C, Acosta LG, Rigoutsos I, Tamayo G, Green BD, Chang C, Rubin EM, Mathur EJ, Robertson DE, Hugenholtz P, Leadbetter JR (2007) Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450:560–565
Weber CF, Stubner S, Conrad R (2001) Bacterial populations colonizing and degrading rice straw in anoxic paddy soil. Appl Environ Microbiol 67:1318–1327
Wongwilaiwalin S, Rattanachomsri U, Laothanachareon T, Eurwilaichitr L, Igarashi Y, Champreda V (2010) Analysis of a thermophilic lignocellulose degrading microbial consortium and multi-species lignocellulolytic enzyme system. Enzym Microb Technol 47:283–290
Zhang Q, He J, Tian M, Maoa Z, Tang L, Zhang J, Zhang H (2011) Enhancement of methane production from cassava residues by biological pretreatment using a constructed microbial consortium. Bioresour Technol 102:8899–8906
Zhu H, Qu F, Zhu LH (1993) Isolation of genomic DNAs from plants, fungi and bacteria using benzyl chloride. Nucleic Acids Res 21:5278–5280
Zuroff TR, Curtis WR (2012) Developing symbiotic consortia for lignocellulosic biofuel production. Appl Microbiol Biotechnol 93:1423–1435
Acknowledgments
This project is under the JSPS-BIOTEC collaborative project supported by a research grant from the National Science and Technology Development Agency (P-11-00926). Proofreading and comments by Dr. Philip Shaw and Dr. Varadom Charoensawan are appreciated.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wongwilaiwalin, S., Laothanachareon, T., Mhuantong, W. et al. Comparative metagenomic analysis of microcosm structures and lignocellulolytic enzyme systems of symbiotic biomass-degrading consortia. Appl Microbiol Biotechnol 97, 8941–8954 (2013). https://doi.org/10.1007/s00253-013-4699-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-013-4699-y