Abstract
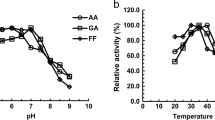
The function of three Corynebacterium glutamicum shikimate dehydrogenase homologues, designated as qsuD (cgR_0495), cgR_1216, and aroE (cgR_1677), was investigated. A disruptant of aroE required shikimate for growth, whereas a qsuD-deficient strain did not grow in medium supplemented with either quinate or shikimate as sole carbon sources. There was no discernible difference in growth rate between wild-type and a cgR_1216-deficient strain. Enzymatic assays showed that AroE both reduced 3-dehydroshikimate, using NADPH as cofactor, and oxidized shikimate, the reverse reaction, using NADP+ as cofactor. The reduction reaction was ten times faster than the oxidation. QsuD reduced 3-dehydroquinate using NADH and oxidized quinate using NAD+ as cofactor. Different from the other two homologues, the product of cgR_1216 displayed considerably lower enzyme activity for both the reduction and the oxidation. The catalytic reaction of QsuD and AroE was highly susceptible to pH. Furthermore, reduction of 3-dehydroshikimate by AroE was inhibited by high concentrations of shikimate, but neither quinate nor aromatic amino acids had any effect on the reaction. Expression of qsuD mRNA was strongly enhanced in the presence of shikimate, whereas that of cgR_1216 and aroE decreased. We conclude that while AroE is the main catalyst for shikimate production in the shikimate pathway, QsuD is essential for quinate/shikimate utilization.





Similar content being viewed by others
References
Anton IA, Coggins JR (1988) Sequencing and overexpression of the Escherichia coli aroE gene encoding shikimate dehydrogenase. Biochem J 249:319–326
Balinsky D, Dennis AW, Cleland WW (1971) Kinetic and isotope-exchange studies on shikimate dehydrogenase from Pisum sativum. Biochemistry 10:1947–1952
Benach J, Lee I, Edstrom W, Kuzin AP, Chiang Y, Acton TB, Montelione GT, Hunt JF (2003) The 2.3-Å crystal structure of the shikimate 5-dehydrogenase orthologue YdiB from Escherichia coli suggests a novel catalytic environment for an NAD-dependent dehydrogenase. J Biol Chem 278:19176–19182
Bongaerts J, Krämer M, Müller U, Raeven L, Wubbolts M (2001) Metabolic engineering for microbial production of aromatic amino acids and derived compounds. Metab Eng 3:289–300
Bonner CA, Jensen RA (1994) Cloning of cDNA encoding the bifunctional dehydroquinase.shikimate dehydrogenase of aromatic-amino-acid biosynthesis in Nicotiana tabacum. Biochem J 302:11–14
Chaudhuri S, Coggins JR (1985) The purification of shikimate dehydrogenase from Escherichia coli. Biochem J 226:217–223
Dell KA, Frost JW (1993) Identification and removal of impediments to biocatalytic synthesis of aromatics from d-glucose: rate-limiting enzymes in the common pathway of aromatic amino acid biosynthesis. J Am Chem Soc 115:11581–11589
Duncan K, Edwards RM, Coggins JR (1987) The pentafunctional arom enzyme of Saccharomyces cerevisiae is a mosaic of monofunctional domains. Biochem J 246:375–386
Ehira S, Ogino H, Teramoto H, Inui M, Yukawa H (2009) Regulation of quinone oxidoreductase by the redox-sensing transcriptional regulator QorR in Corynebacterium glutamicum. J Biol Chem 284:16736–16742
Follmann M, Ochrombel I, Krämer R, Trötschel C, Poetsch A, Rückert C, Hüser A, Persicke M, Seiferling D, Kalinowski J, Marin K (2009) Functional genomics of pH homeostasis in Corynebacterium glutamicum revealed novel links between pH response, oxidative stress, iron homeostasis and methionine synthesis. BMC Genomics 10:621
Fonseca IO, Magalhaes ML, Oliveira JS, Silva RG, Mendes MA, Palma MS, Santos DS, Basso LA (2006) Functional shikimate dehydrogenase from Mycobacterium tuberculosis H37Rv: purification and characterization. Protein Expr Purif 46:429–437
Fonseca IO, Silva RG, Fernandes CL, de Souza ON, Basso LA, Santos DS (2007) Kinetic and chemical mechanisms of shikimate dehydrogenase from Mycobacterium tuberculosis. Arch Biochem Biophys 457:123–133
Han C, Wang L, Yu K, Chen L, Hu L, Chen K, Jiang H, Shen X (2006) Biochemical characterization and inhibitor discovery of shikimate dehydrogenase from Helicobacter pylori. FEBS J 273:4682–4692
Hermann T (2003) Industrial production of amino acids by coryneform bacteria. J Biotechnol 104:155–172
Herrmann KM, Weaver LM (1999) The shikimate pathway. Annu Rev Plant Physiol Plant Mol Biol 50:473–503
Ikeda M, Nakanishi K, Kino K, Katsumata R (1994) Fermentative production of tryptophan by a stable recombinant strain of Corynebacterium glutamicum with a modified serine-biosynthetic pathway. Biosci Biotechnol Biochem 58:674–678
Ikeda M, Katsumata R (1999) Hyperproduction of tryptophan by Corynebacterium glutamicum with the modified pentose phosphate pathway. Appl Environ Microbiol 65:2497–2502
Inui M, Murakami S, Okino S, Kawaguchi H, Vertès AA, Yukawa H (2004) Metabolic analysis of Corynebacterium glutamicum during lactate and succinate productions under oxygen deprivation conditions. J Mol Microbiol Biotechnol 7:182–196
Kikuchi Y, Tsujimoto K, Kurahashi O (1997) Mutational analysis of the feedback sites of phenylalanine-sensitive 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase of Escherichia coli. Appl Environ Microbiol 63:761–762
Kishore GM, Shah DM (1988) Amino acid biosynthesis inhibitors as herbicides. Annu Rev Biochem 57:627–663
Knaggs AR (2003) The biosynthesis of shikimate metabolites. Nat Prod Rep 20:119–136
Koma D, Yamanaka H, Moriyoshi K, Ohmoto T, Sakai K (2012) Production of aromatic compounds by metabolically engineered Escherichia coli with an expanded shikimate pathway. Appl Environ Microbiol 78:6203–6216
Lumsden J, Coggins JR (1977) The subunit structure of the arom multienzyme complex of Neurospora crassa. A possible pentafunctional polypeptide chain. Biochem J 161:599–607
Maddocks SE, Oyston PC (2008) Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154:3609–3623
Michel G, Roszak AW, Sauvé V, Maclean J, Matte A, Coggins JR, Cygler M, Lapthorn AJ (2003) Structures of shikimate dehydrogenase AroE and its Paralog YdiB. A common structural framework for different activities. J Biol Chem 278:19463–19472
Ray JM, Yanofsky C, Bauerle R (1988) Mutational analysis of the catalytic and feedback sites of the tryptophan-sensitive 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase of Escherichia coli. J Bacteriol 170:5500–5506
Richards TA, Dacks JB, Campbell SA, Blanchard JL, Foster PG, McLeod R, Roberts CW (2006) Evolutionary origins of the eukaryotic shikimate pathway: gene fusions, horizontal gene transfer, and endosymbiotic replacements. Eukaryot Cell 5:1517–1531
Schoepe J, Niefind K, Schomburg D (2008) 1.6 angstroms structure of an NAD+-dependent quinate dehydrogenase from Corynebacterium glutamicum. Acta Crystallogr D: Biol Crystallogr 64:803–809
Singh S, Korolev S, Koroleva O, Zarembinski T, Collart F, Joachimiak A, Christendat D (2005) Crystal structure of a novel shikimate dehydrogenase from Haemophilus influenzae. J Biol Chem 280:17101–17108
Singh S, Stavrinides J, Christendat D, Guttman DS (2008) A phylogenomic analysis of the shikimate dehydrogenases reveals broadscale functional diversification and identifies one functionally distinct subclass. Mol Biol Evol 25:2221–2232
Tanaka Y, Okai N, Teramoto H, Inui M, Yukawa H (2008) Regulation of the expression of phosphoenolpyruvate: Carbohydrate phosphotransferase system (PTS) genes in Corynebacterium glutamicum R. Microbiology 154:264–274
Teramoto H, Inui M, Yukawa H (2009) Regulation of expression of genes involved in quinate and shikimate utilization in Corynebacterium glutamicum. Appl Environ Microbiol 75:3461–3468
Weaver LM, Herrmann KM (1990) Cloning of an aroF allele encoding a tyrosine-insensitive 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase. J Bacteriol 172:6581–6584
Ye S, Von Delft F, Brooun A, Knuth MW, Swanson RV, McRee DE (2003) The crystal structure of shikimate dehydrogenase (AroE) reveals a unique NADPH binding mode. J Bacteriol 185:4144–4151
Yu TW, Müller R, Müller M, Zhang X, Draeger G, Kim CG, Leistner E, Floss HG (2001) Mutational analysis and reconstituted expression of the biosynthetic genes involved in the formation of 3-amino-5-hydroxybenzoic acid, the starter unit of rifamycin biosynthesis in Amycolatopsis mediterranei S699. J Biol Chem 276:12546–12555
Yukawa H, Omumasaba CA, Nonaka H, Kós P, Okai N, Suzuki N, Suda M, Tsuge Y, Watanabe J, Ikeda Y, Vertès AA, Inui M (2007) Comparative analysis of the Corynebacterium glutamicum group and complete genome sequence of strain R. Microbiology 153:1042–1058
Zhang X, Zhang S, Hao F, Lai X, Yu H, Huang Y, Wang H (2005) Expression, purification and properties of shikimate dehydrogenase from Mycobacterium tuberculosis. J Biochem Mol Biol 38:624–631
Acknowledgments
We thank Crispinus A. Omumasaba (RITE) for critical reading of the manuscript. This work was partially supported by a grant from the New Energy and Industrial Technology Development Organization (NEDO).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 187 kb)
Rights and permissions
About this article
Cite this article
Kubota, T., Tanaka, Y., Hiraga, K. et al. Characterization of shikimate dehydrogenase homologues of Corynebacterium glutamicum . Appl Microbiol Biotechnol 97, 8139–8149 (2013). https://doi.org/10.1007/s00253-012-4659-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-012-4659-y