Abstract
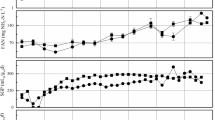
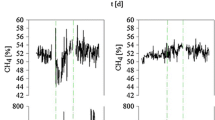
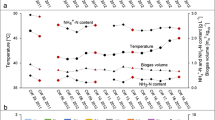
Laboratory biogas reactors were operated under various conditions using maize silage, chicken manure, or distillers grains as substrate. In addition to the standard process parameters, the hydrogen and carbon stable isotopic composition of biogas was analyzed to estimate the predominant methanogenic pathways as a potential process control tool. The isotopic fingerprinting technique was evaluated by parallel analysis of mcrA genes and their transcripts to study the diversity and activity of methanogens. The dominant hydrogenotrophs were Methanomicrobiales, while aceticlastic methanogens were represented by Methanosaeta and Methanosarcina at low and high organic loading rates, respectively. Major changes in the relative abundance of mcrA transcripts were observed compared to the results obtained from DNA level. In agreement with the molecular results, the isotope data suggested the predominance of the hydrogenotrophic pathway in one reactor fed with chicken manure, while both pathways were important in the other reactors. Short-term changes in the isotopic composition were followed, and a significant change in isotope values was observed after feeding a reactor digesting maize silage. This ability of stable isotope fingerprinting to follow short-term activity changes shows potential for indicating process failures and makes it a promising technology for process control.




Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410
Angelidaki I, Ellegaard L, Ahring BK (1993) A mathematical model for dynamic simulation of anaerobic digestion of complex substrates: focusing on ammonia inhibition. Biotechnol Bioeng 42(2):159–66. doi:10.1002/bit.260420203
Bauer C, Korthals M, Gronauer A, Lebuhn M (2008) Methanogens in biogas production from renewable resources—a novel molecular population analysis approach. Water Sci Technol 58(7):1433–1439. doi:10.2166/Wst.2008.514
Blume F, Bergmann I, Nettmann E, Schelle H, Rehde G, Mundt K, Klocke M (2010) Methanogenic population dynamics during semi-continuous biogas fermentation and acidification by overloading. J Appl Microbiol 109(2):441–450. doi:10.1111/j.1365-2672.2010.04682.x
Briones A, Raskin L (2003) Diversity and dynamics of microbial communities in engineered environments and their implications for process stability. Curr Opin Biotechnol 14(3):270–276. doi:10.1016/S0958-1669(03)00065-X
Conrad R (2005) Quantification of methanogenic pathways using stable carbon isotopic signatures: a review and a proposal. Org Geochem 36(5):739–752. doi:10.1016/j.orggeochem.2004.09.006
Demirel B, Scherer P (2008) The roles of acetotrophic and hydrogenotrophic methanogens during anaerobic conversion of biomass to methane: a review. Rev Environ Sci Biotechnol 7:173–190
Feisthauer S, Siegert M, Seidel M, Richnow HH, Zengler K, Grundger F, Kruger M (2010) Isotopic fingerprinting of methane and CO2 formation from aliphatic and aromatic hydrocarbons. Org Geochem 41(5):482–490. doi:10.1016/j.orggeochem.2010.01.003
Fernandez A, Huang SY, Seston S, Xing J, Hickey R, Criddle C, Tiedje J (1999) How stable is stable? Function versus community composition. Appl Environ Microbiol 65(8):3697–3704
Goberna M, Schoen MA, Sperl D, Wett B, Insam H (2010) Mesophilic and thermophilic co-fermentation of cattle excreta and olive mill wastes in pilot anaerobic digesters. Biomass Bioenergy 34(3):340–346. doi:10.1016/j.biombioe.2009.11.005
Hanreich A, Heyer R, Benndorf D, Rapp E, Pioch M, Reichl U, Klocke M (2012) Metaproteome analysis to determine the metabolically active part of a thermophilic microbial community producing biogas from agricultural biomass. Can J Microbiol 58(7):917–922. doi:10.1139/W2012-058
Keppler F, Laukenmann S, Rinne J, Heuwinkel H, Greule M, Whiticar M, Lelieveld J (2010) Measurements of 13C/12C methane from anaerobic digesters: Comparison of optical spectrometry with continuous-flow isotope ratio mass spectrometry. Environ Sci Technol 44(13):5067–5073. doi:10.1021/Es100460d
Krzycki JA, Kenealy WR, Deniro MJ, Zeikus JG (1987) Stable carbon isotope fractionation by Methanosarcina barkeri during methanogenesis from acetate, methanol, or carbon dioxide–hydrogen. Appl Environ Microbiol 53(10):2597–2599
Laukenmann S, Polag D, Heuwinkel H, Greule M, Gronauer A, Lelieveld J, Keppler F (2010) Identification of methanogenic pathways in anaerobic digesters using stable carbon isotopes. Eng Life Sci 10(6):509–514. doi:10.1002/elsc.201000074
Leclerc M, Delgenes JP, Godon JJ (2004) Diversity of the archaeal community in 44 anaerobic digesters as determined by single strand conformation polymorphism analysis and 16S rDNA sequencing. Environ Microbiol 6(8):809–819. doi:10.1111/j.1462-2920.2004.00616.x
Li TL, Mazeas L, Sghir A, Leblon G, Bouchez T (2009) Insights into networks of functional microbes catalysing methanization of cellulose under mesophilic conditions. Environ Microbiol 11(4):889–904. doi:10.1111/j.1462-2920.2008.01810.x
Liu FH, Wang SB, Zhang JS, Zhang J, Yan X, Zhou HK, Zhao GP, Zhou ZH (2009) The structure of the bacterial and archaeal community in a biogas digester as revealed by denaturing gradient gel electrophoresis and 16S rDNA sequencing analysis. J Appl Microbiol 106(3):952–66. doi:10.1111/j.1365-2672.2008.04064.x
Liu YC, Whitman WB (2008) Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Incredible anaerobes. From Physiology to Genomics to Fuels 1125:171–189. doi:10.1196/annals.1419.019
Luton PE, Wayne JM, Sharp RJ, Riley PW (2002) The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology-SGM 148:3521–3530
Meckenstock RU, Morasch B, Griebler C, Richnow HH (2004) Stable isotope fractionation analysis as a tool to monitor biodegradation in contaminated acquifers. J Contam Hydrol 75(3–4):215–55. doi:10.1016/j.jconhyd.2004.06.003
Munk B, Bauer C, Gronauer A, Lebuhn M (2010) Population dynamics of methanogens during acidification of biogas fermenters fed with maize silage. Eng Life Sci 10(8):496–508
Nettmann E, Bergmann I, Mundt K, Linke B, Klocke M (2008) Archaea diversity within a commercial biogas plant utilizing herbal biomass determined by 16S rDNA and mcrA analysis. J Appl Microbiol 105(6):1835–1850. doi:10.1111/j.1365-2672.2008.03949.x
Nikolausz M, Kappelmeyer U, Szekely A, Rusznyak A, Marialigeti K, Kastner M (2008) Diurnal redox fluctuation and microbial activity in the rhizosphere of wetland plants. Eur J Soil Biol 44(3):324–333. doi:10.1016/j.ejsobi.2008.01.003
O’Leary MH (1988) Carbon isotopes in photosynthesis. BioScience 38(5):328–336
Qu X, Mazeas L, Vavilin VA, Epissard J, Lemunier M, Mouchel JM, He PJ, Bouchez T (2009a) Combined monitoring of changes in delta13CH4 and archaeal community structure during mesophilic methanization of municipal solid waste. FEMS Microbiol Ecol 68(2):236–245. doi:10.1111/j.1574-6941.2009.00661.x
Qu X, Vavilin VA, Mazeas L, Lemunier M, Duquennoi C, He PJ, Bouchez T (2009b) Anaerobic biodegradation of cellulosic material: batch experiments and modelling based on isotopic data and focusing on aceticlastic and non-aceticlastic methanogenesis. Waste Manag 29(6):1828–1837. doi:10.1016/j.wasman.2008.12.008
Rastogi G, Ranade DR, Yeole TY, Patole MS, Shouche YS (2008) Investigation of methanogen population structure in biogas reactor by molecular characterization of methyl-coenzyme M reductase A (mcrA) genes. Bioresour Technol 99(13):5317–5326. doi:10.1016/j.biortech.2007.11.024
Schluter A, Bekel T, Diaz NN, Dondrup M, Eichenlaub R, Gartemann KH, Krahn I, Krause L, Kromeke H, Kruse O, Mussgnug JH, Neuweger H, Niehaus K, Puhler A, Runte KJ, Szczepanowski R, Tauch A, Tilker A, Viehover P, Goesmann A (2008) The metagenome of a biogas-producing microbial community of a production-scale biogas plant fermenter analysed by the 454-pyrosequencing technology. J Biotech 136(1–2):77–90. doi:10.1016/j.jbiotec.2008.05.008
Schnürer A, Nordberg A (2008) Ammonia, a selective agent for methane production by syntrophic acetate oxidation at mesophilic temperature. Water Sci Technol 57(5):735–740. doi:10.2166/Wst.2008.097
Schnürer A, Zellner G, Svensson BH (1999) Mesophilic syntrophic acetate oxidation during methane formation in biogas reactors. FEMS Microbiol Ecol 29(3):249–261
Steinberg LM, Regan JM (2008) Phylogenetic comparison of the methanogenic communities from an acidic, oligotrophic fen and an anaerobic digester treating municipal wastewater sludge. Appl Environ Microbiol 74(21):6663–6671. doi:10.1128/Aem.00553-08
Sugimoto A, Wada E (1995) Hydrogen isotopic composition of bacterial methane—CO2/H2 reduction and acetate fermentation. Geochim Cosmochim Acta 59(7):1329–1337
Talbot G, Topp E, Palin MF, Masse DI (2008) Evaluation of molecular methods used for establishing the interactions and functions of microorganisms in anaerobic bioreactors. Water Res 42(3):513–537. doi:10.1016/j.watres.2007.08.003
Vavilin VA, Qu X, Mazeas L, Lemunier M, Duquennoi C, He PJ, Bouchez T (2008) Methanosarcina as the dominant aceticlastic methanogens during mesophilic anaerobic digestion of putrescible waste. Antonie van Leeuwenhoek 94(4):593–605. doi:10.1007/s10482-008-9279-2
Whiticar MJ (1999) Carbon and hydrogen isotope systematics of bacterial formation and oxidation of methane. Chem Geol 161(1–3):291–314
Whiticar MJ, Faber E, Schoell M (1986) Biogenic methane formation in marine and fresh-water environments—CO2 reduction vs acetate fermentation isotope evidence. Geochim Cosmochim Acta 50(5):693–709
Yang ST, Tang IC (1991) Methanogenesis from lactate by a coculture of Clostridium formicoaceticum and Methanosarcina mazei. Appl Microbiol Biotechnol 35(1):119–123
Zakrzewski M, Goesmann A, Jaenicke S, Junemann S, Eikmeyer F, Szczepanowski R, Abu Al-Soud W, Sorensen S, Puhler A, Schluter A (2012) Profiling of the metabolically active community from a production-scale biogas plant by means of high-throughput metatranscriptome sequencing. J Biotech 158(4):248–258. doi:10.1016/j.jbiotec.2012.01.020
Ziganshin AM, Schmidt T, Scholwin F, Il'inskaya ON, Harms H, Kleinsteuber S (2011) Bacteria and Archaea involved in anaerobic digestion of distillers grains with solubles. Appl Microbiol Biotechnol 89(6):2039–2052
Acknowledgments
The study was supported by the Initiative and Networking Fund of the Helmholtz Association. We would like to acknowledge Petra Blümel (UFZ—Department Catchment Hydrology) for providing hydrogen isotope data of the tap water in Leipzig. We would like to thank Katrin Strach and Robin Bujak (DBFZ) for their help in operating the biogas reactors and performing analytics.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 603 kb)
Rights and permissions
About this article
Cite this article
Nikolausz, M., Walter, R.F.H., Sträuber, H. et al. Evaluation of stable isotope fingerprinting techniques for the assessment of the predominant methanogenic pathways in anaerobic digesters. Appl Microbiol Biotechnol 97, 2251–2262 (2013). https://doi.org/10.1007/s00253-012-4657-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-012-4657-0