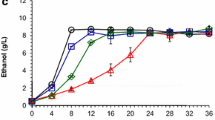
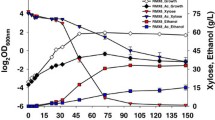
Abstract
Furfural from lignocellulosic hydrolysates is the prevalent inhibitor to microorganisms during cellulosic ethanol production, but the molecular mechanisms of tolerance to this inhibitor in Zymomonas mobilis are still unclear. In this study, genome-wide transcriptional responses to furfural were investigated in Z. mobilis using microarray analysis. We found that 433 genes were differentially expressed in response to furfural. Furfural up- or down-regulated genes related to cell wall/membrane biogenesis, metabolism, and transcription. However, furfural has a subtle negative effect on Entner–Doudoroff pathway mRNAs. Our results revealed that furfural had effects on multiple aspects of cellular metabolism at the transcriptional level and that membrane might play important roles in response to furfural. This research has provided insights into the molecular response to furfural in Z. mobilis, and it will be helpful to construct more furfural-resistant strains for cellulosic ethanol production.





Similar content being viewed by others
References
Allen SA, Clark W, McCaffery JM, Cai Z, Lanctot A, Slininger PJ, Liu ZL, Gorsich SW (2010) Furfural induces reactive oxygen species accumulation and cellular damage in Saccharomyces cerevisiae. Biotechnol Biofuels 3(1):2–12
Almeida JRM, Modig T, Petersson A, Hahn-Hagerdal B, Liden G, Gorwa-Grauslund MF (2007) Increased tolerance and conversion of inhibitors in lignocellulosic hydrolysates by Saccharomyces cerevisiae. J Chem Tech Biotech 82(4):340–349
Almeida JRM, Roder A, Modig T, Laddan B, Liden G, Gorwa-Grauslund MF (2008) NADH- vs. NADPH-coupled reduction of 5-hydroxymethyl furfural (HMF) and its implications on product distribution in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 78:839–945
Barciszewski J, Siboska GE, Pedersen BO, Clark BFC, Ratten SIS (1997) A mechanism for the in vivo formation of N-6-furfuryladenine, kinetin, as a secondary oxidative damage product of DNA. FEBS Lett 414:457–460
Brissette JL, Russel M, Weiner L, Model P (1990) Phage shock protein, a stress protein of Escherichia coli. P Natl Acad Sci USA 87:862–866
Chomczynski P (1993) A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. BioTechniques 15:532–537
Deanda K, Zhang M, Eddy C, Picataggio S (1996) Development of an arabinose-fermenting Zymomonas mobilis strain by metabolic pathway engineering. Appl Environ Microbiol 62(12):4465–4470
Franden MA, Pienkos PT, Zhang M (2009) Development of a high-throughput method to evaluate the impact of inhibitory compounds from lignocellulosic hydrolysates on the growth of Zymomonas mobilis. J Biotechnol 144(4):259–267
Goodman AE, Rogers PL, Skotnicki ML (1982) Minimal medium for isolation of auxotrophic Zymomonas mutants. Appl Environ Microbiol 44(2):496–498
Gorsich SW, Dien BS, Nichols NN, Slininger PJ, Liu ZL, Skory CD (2006) Tolerance to furfural-induced stress is associated with pentose phosphate pathway genes ZWF1, GND1, RPE1, and TKL1 in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 71:339–349
Gutiérrez T, Ingram LO, Preston JF (2006) Purification and characterization of a furfural reductase (FFR) from Escherichia coli strain LY01-an enzyme important in the detoxification of furfural during ethanol production. J Biotechnol 121:154–164
Gutierrez-Padilla MGD, Karim MN (2005) Influence of furfural on the recombinant Zymomonas mobilis strain CP4 (pZB5) for ethanol production. J Am Sci 1(1):24–27
Horbach S, Neuss B, Sahm H (1991) Effect of azasqualene on hopanoid biosynthesis and ethanol tolerance of Zymomonas mobilis. FEMS Microbiol Lett 79(2–3):347–350
Hung SP, Baldi P, Hatfield GW (2002) Global gene expression profiling in Escherichia coli K12. The effects of leucine-responsive regulatory protein. J Biol Chem 277:40309–40323
Hussein R, Lim HN (2011) Disruption of small RNA signaling caused by competition for Hfq. Proc Natl Acad Sci USA 108(3):1110–1115
Huvet M, Toni T, Sheng X, Thorne T, Jovanovic G, Engl C, Buck M, Pinney JW, Stumpf MPH (2011) The evolution of the phage shock protein response system: interplay between protein function, genomic organization, and system function. Mol Biol Evol 28(3):1141–1155
Jeffries TW (2005) Ethanol fermentation on the move. Nat Biotechnol 2005(23):40–41
Joly N, Engl C, Jovanovic G, Huvet M, Toni T, Sheng X, Stumpf MPH, Buck M (2010) Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol Rev 34(5):797–827
Kalnenieks U (2006) Physiology of Zymomonas mobilis: some unanswered questions. Adv Microb Physiol 51:73–117
Kerr AL, Jeon YJ, Svenson CJ, Rogers PL, Neilan BA (2010) DNA restriction–modification systems in the ethanologen, Zymomonas mobilis ZM4. Appl Microbiol Biotech 89(3):761–769
Khan QA, Shamsi FA, Hadi SM (1995) Mutagenicity of furfural in plasmid DNA. Cancer Lett 89:95–99
Koopman F, Wierckx N, de Winde JH, Ruijssenaars HJ (2010) Identification and characterization of the furfural and 5-(hydroxymethyl)furfural degradation pathways of Cupriavidus basilensis HMF14. Proc Natl Acad Sci USA 107(11):4919–4924
Kouvelis VN, Saunders E, Brettin TS, Bruce D, Detter C, Han C, Typas MA, Pappas KM (2009) Complete genome sequence of the ethanol producer Zymomonas mobilis NCIMB 11163. J Bacteriol 191(22):7140–7141
Kouvelis VN, Davenport KW, Brettin TS, Bruce D, Detter C, Han C, Nolan M, Tapia R, Damoulaki A, Kyrpides NC, Typas MA, Pappas KM (2011) Genome sequence of the ethanol-producing Zymomonas mobilis subsp. pomaceae lectotype ATCC 29192. J Bacteriol 193(18):5049–5050
Lin F-M, Qiao B, Yuan Y-J (2009a) Comparative proteomic analysis of tolerance and adaptation of ethanologenic Saccharomyces cerevisiae to furfural, a lignocellulosic inhibitory compound. Appl Environ Microbiol 75(11):3765–3776
Lin F-M, Tan Y, Yuan Y-J (2009b) Temporal quantitative proteomics of Saccharomyces cerevisiae in response to a nonlethal concentration of furfural. Proteomics 9:5471–5483
Linger JG, Adney WS, Darzins A (2010) Heterologous expression and extracellular secretion of cellulolytic enzymes by Zymomonas mobilis. Appl Environ Microbiol 76(19):6360–6369
Liu ZL (2006) Genomic adaptation of ethanologenic yeast to biomass conversion inhibitors. Appl Microbiol Biotechnol 73:27–36
Liu ZL, Moon J, Andersh BJ, Slininger PJ, Weber S (2008) Multiple gene-mediated NAD(P)H-dependent aldehyde reduction is a mechanism of in situ detoxification of furfural and 5-hydroxyfurfural by Saccharomyces cerevisiae. Appl Microbiol Biotechnol 81:743–753
Miller EN, Jarboe LR, Yomano LP, York SW, Shanmugam KT, Ingram LO (2009) Silencing of NADPH-dependent oxidoreductase genes (yqhD and dkgA) in furfural-resistant ethanologenic Escherichia coli. Appl Enviro Microbiol 75(13):4315–4323
Miller EN, Turner PC, Jarboe LR, Ingram LO (2010) Genetic changes that increase 5-hydroxymethyl furfural resistance in ethanol-producing Escherichia coli LY180. Biotchnol Lett 32(5):661–667
Mills TY, Sandoval NR, Gill RT (2009) Cellulosic hydrolysate toxicity and tolerance mechanisms in Escherichia coli. Biotchnol Biofuels 2:26–37
Palmqvist E, Hahn-Hagerdal B (2000) Fermentation of lignocellulosic hydrolysates. II: inhibitors and mechanisms of inhibition. Biores Technol 74:25–33
Pappas KM, Kouvelis VN, Saunders E, Brettin TS, Bruce D, Detter C, Balakireva M, Han C, Savvakis G, Kyrpides NC, Typas MA (2011) Genome sequence of the ethanol-producing Zymomonas mobilis subsp. mobilis lectotype ATCC 10988. J Bacteriol 193(18):5051–5052
Perzl M, Reipen IG, Schmitz S, Poralla K, Sahm H, Sprenger GA, Elmar L (1998) Cloning of conserved genes from Zymomonas mobilis and Bradyrhizobium japonicum that function in the biosynthesis of hopanoid lipids. Biochimica et Biophysica Acta (BBA) - Lipids and Lipid Metabolism 1393(1):108–118
Pienkos P, Zhang M (2009) Role of pretreatment and conditioning processes on toxicity of lignocellulosic biomass hydrolysates. Cellulose 16:743–762
Rabhi M, Espeli O, Schwartz A, Cayrol B, Rahmouni AR, Arluison V, Boudvillain M (2011) The Sm-like RNA chaperone Hfq mediates transcription antitermination at Rho-dependent terminators. Embo J 30(14):2805–2816
Ranatunga T, Jervis J, Helm R, McMillan J, Hatzis C (1997) Identification of inhibitory components toxic toward Zymomonas mobilis CP4(pZB5) xylose fermentation. Applied Biochem Biotech 67(3):185–198
Schmehl M, Jahn A, Vilsendorf AMz, Hennecke S, Masepohl B, Schuppler M, Marxer M, Oelze J, Klipp W (1993) Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: a putative membrane complex involved in electron transport to nitrogenase. Mol Gen Genet 241:602–615
Seo JS, Chong H, Park HS, Yoon KO, Jung C, Kim JJ, Hong JH, Kim H, Kim JH, Kil JI, Park CJ, Oh HM, Lee JS, Jin SJ, Um HW, Lee HJ, Oh SJ, Kim JY, Kang HL, Lee SY, Lee KJ, Kang HS (2005) The genome sequence of the ethanologenic bacterium Zymomonas mobilis ZM4. Nat Biotechnol 23(1):63–68
Sootsuwan K, Lertwattanasakul N, Thanonkeo P, Matsushita K, Yamada M (2008) Analysis of the respiratory chain in ethanologenic Zymomonas mobilis with a cyanide-resistant bd-type ubiquinol oxidase as the only terminal oxidase and its possible physiological roles. J Mol Microbiol Biotechnol 14(4):163–175
Storz G, Vogel J, Wassarman Karen M (2011) Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell 43(6):880–891
Swings J, Deley J (1977) Biology of Zymomonas. Bacterial Rev 41(1):1–46
Tani TH, Khodursky A, Blumenthal RM, Brown PO, Matthews RG (2002) Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc Natl Acad Sci USA 99:13471–13476
Vogel J, Luisi BF (2011) Hfq and its constellation of RNA. Nat Rev Micro 9(8):578–589
Wang X, Miller EN, Yomano LP, Zhang X, Shanmugam KT, Ingram LO (2011) Increased furfural tolerance due to overexpression of NADH-dependent oxidoreductase FucO in Escherichia coli strains engineered for the production of ethanol and lactate. Appl Environ Microbiol 77(15):5132–5140
Welander PV, Hunter RC, Zhang L, Sessions AL, Summons RE, Newman DK (2009) Hopanoids play a role in membrane integrity and pH homeostasis in Rhodopseudomonas palustris TIE-1. J Bacteriol 191(19):6145–6156
Widiastuti H, Kim JY, Selvarasu S, Karimi IA, Kim H, Seo JS, Lee DY (2011) Genome-scale modeling and in silico analysis of ethanologenic bacteria Zymomonas mobilis. Biotechnol Bioeng 108(3):655–65
Yang S, Tschaplinski TJ, Engle NL, Carroll SL, Martin SL, Davison BH, Palumbo AV, Rodriguez M Jr, Brown SD (2009) Transcriptomic and metabolomic profiling of Zymomonas mobilis during aerobic and anaerobic fermentations. BMC Genomics 10:34
Yang S, Pelletier DA, Lu TY, Brown SD (2010) The Zymomonas mobilis regulator hfq contributes to tolerance against multiple lignocellulosic pretreatment inhibitors. BMC Microbiol 10:135
Zaldivar J, Martinez A, Ingram LO (1999) Effect of selected aldehydes on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol Bioeng 65:24–33
Zaldivar J, Martinez A, Ingram LO (2000) Effect of alcohol compounds found in hemicellulose hydrolysate on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol Bioeng 68:524–530
Zhang M, Eddy C, Deanda K, Finkelstein M, Picataggio S (1995) Metabolic engineering of a pentose metabolism pathway in ethanologenic Zymomonas mobilis. Science 267(5195):240–243
Zhang J, Zhu Z, Wang X, Wang N, Wang W, Bao J (2010) Biodetoxification of toxins generated from lignocellulose pretreatment using a newly isolated fungus, Amorphotheca resinae ZN1, and the consequent ethanol fermentation. Biotechnol Biofuels 3(1):26–41
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 31000028), Sichuan Key Technology R&D Program (Grant No. 2009NZ00045), and Sci-tech Fund Project of Chinese Academy of Agricultural Sciences (2009 and 2011).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 56 kb)
Rights and permissions
About this article
Cite this article
He, Mx., Wu, B., Shui, Zx. et al. Transcriptome profiling of Zymomonas mobilis under furfural stress. Appl Microbiol Biotechnol 95, 189–199 (2012). https://doi.org/10.1007/s00253-012-4155-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-012-4155-4