Abstract
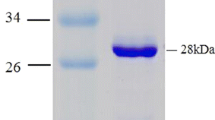
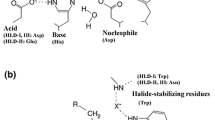
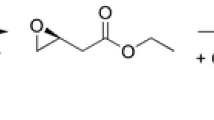
A haloalkane dehalogenase (DppA) from Plesiocystis pacifica SIR-1 was identified by sequence comparison in the NCBI database, cloned, functionally expressed in Escherichia coli, purified, and biochemically characterized. The three-dimensional (3D) structure was determined by X-ray crystallography and has been refined at 1.95 Å resolution to an R-factor of 21.93%. The enzyme is composed of an α/β-hydrolase fold and a cap domain and the overall fold is similar to other known haloalkane dehalogenases. Active site residues were identified as Asp123, His278, and Asp249 and Trp124 and Trp163 as halide-stabilizing residues. DppA, like DhlA from Xanthobacter autotrophicus GJ10, is a member of the haloalkane dehalogenase subfamily HLD-I. As a consequence, these enzymes have in common the relative position of their catalytic residues within the structure and also show some similarities in the substrate specificity. The enzyme shows high preference for 1-bromobutane and does not accept chlorinated alkanes, halo acids, or halo alcohols. It is a monomeric protein with a molecular mass of 32.6 kDa and exhibits maximum activity between 33 and 37°C with a pH optimum between pH 8 and 9. The Km and kcat values for 1-bromobutane were 24.0 mM and 8.08 s−1. Furthermore, from the 3D-structure of DppA, it was found that the enzyme possesses a large and open active site pocket. Docking experiments were performed to explain the experimentally determined substrate preferences.







Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Bidmanova S, Chaloupkova R, Damborsky J, Prokop Z (2010) Development of an enzymatic fiber-optic biosensor for detection of halogenated hydrocarbons. Anal Bioanal Chem 398:1891–1898
Bogdanović X, Hesseler M, Palm GJ, Bornscheuer UT, Hinrichs W (2010) Crystallization and preliminary X-ray diffraction studies of the putative haloalkane dehalogenase DppA from Plesiocystis pacifica SIR-I. Acta Crystallogr F Struct Biol Cryst Commun 66:828–830
Carr PD, Ollis DL (2009) Alpha/beta hydrolase fold: an update. Protein Pept Lett 16:1137–1148
Chaloupkova R, Sykorova J, Prokop Z, Jesenska A, Monincova M, Pavlova M, Tsuda M, Nagata Y, Damborsky J (2003) Modification of activity and specificity of haloalkane dehalogenase from Sphingomonas paucimobilis UT26 by engineering of its entrance tunnel. J Biol Chem 278:52622–52628
Chovancova E, Kosinski J, Bujnicki JM, Damborsky J (2007) Phylogenetic analysis of haloalkane dehalogenases. Proteins 67:305–316
Collaborative Computational Project N (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50:760–763
Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16:10881–10890
Damborsky J, Rorije E, Jesenska A, Nagata Y, Klopman G, Peijnenburg WJGM (2001) Structure-specificity relationships for haloalkane dehalogenases. Environ Toxicol Chem 20:2681–2689
Damborsky J, Chaloupkova R, Pavlova M, Chovancova E, Brezovsky J (2010) Structure-function relationships and engineering of haloalkane dehalogenases. In: Timmis KN (ed) Handbook of hydrocarbon and lipid microbiology. Springer Berlin Heidelberg, pp 1081–1098
Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB 3rd, Snoeyink J, Richardson JS, Richardson DC (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35(Web Server issue):W375–W383
Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J (2006) CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res 34(Web Server issue):W116–W118
Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60(Pt 12 Pt 1):2126–2132
Franken SM, Rozeboom HJ, Kalk KH, Dijkstra BW (1991) Crystal-structure of haloalkane dehalogenase—an enzyme to detoxify halogenated alkanes. EMBO J 10:1297–1302
Gouet P, Courcelle E, Stuart DI, Metoz F (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15:305–308
Holm L, Rosenstrom P (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res 38(Web Server issue):W545–W549
Holmquist M (2000) Alpha/beta-hydrolase fold enzymes: structures, functions and mechanisms. Curr Protein Pept Sci 1:209–235
Iwasaki I, Utsumi S, Ozawa T (1952) New colorimetric determination of chloride using mercuric thiocyanate and ferric ion. Bull Chem Soc Jpn 25:226
Janssen DB (2004) Evolving haloalkane dehalogenases. Curr Opin Chem Biol 8:150–159
Janssen DB (2007) Biocatalysis by dehalogenating enzymes. Adv Appl Microbiol 61:233–252
Janssen D, Wijngaard A, Waarde J, Oldenhuis R (1991) In: Hinchee RE, Olfenbuttel RF (eds) On-site bioreclamation, 1st edn. Butterworth-Heinmann, Boston, pp 539–547
Janssen DB, Dinkla IJT, Poelarends GJ, Terpstra P (2005) Bacterial degradation of xenobiotic compounds: evolution and distribution of novel enzyme activities. Environ Microbiol 7:1868–1882
Jesenska A, Pavlova M, Strouhal M, Chaloupkova R, Tseinska I, Monincova M, Prokop Z, Bartos M, Pavlik I, Rychlik I, Möbius P, Nagata Y, Damborsky J (2005) Cloning, biochemical properties, and distribution of mycobacterial haloalkane dehalogenases. Appl Environ Microbiol 71:6736–6745
Jesenska A, Monincova M, Koudelakova T, Hasan K, Chaloupkova R, Prokop Z, Geerlof A, Damborsky J (2009) Biochemical characterization of haloalkane dehalogenases DrbA and DmbC: representatives of novel subfamily. Appl Environ Microbiol 75:5157–5160, AEM 00199-00209
Keuning S, Janssen DB, Witholt B (1985) Purification and characterization of hydrolytic haloalkane dehalogenase from Xanthobacter-autotrophicus Gj10. J Bacteriol 163:635–639
Krooshof GH, Kwant EM, Damborsky J, Koca J, Janssen DB (1997) Repositioning the catalytic triad aspartic acid of haloalkane dehalogenase: effects on stability, kinetics, and structure. Biochemistry 36:9571–9580
Iizuka T, Jojima Y, Fudou R, Hiraishi A, Ahn J-W, Yamanaka S (2003) Plesiocystis pacifica gen. nov., sp. nov., a marine myxobacterium that contains dihydrogenated menaquinone, isolated from the Pacific coasts of Japan. Int J Syst Evol Microbiol 53(Pt 1):189–195
Los GV, Wood K (2007) The HaloTag: a novel technology for cell imaging and protein analysis. Meth Mol Biol 356:195–208
Marek J, Vevodova J, Smatanova IK, Nagata Y, Svensson LA, Newman J, Takagi M, Damborsky J (2000) Crystal structure of the haloalkane dehalogenase from Sphingomonas paucimobilis UT26. Biochemistry 39:14082–14086
Marvanova S, Nagata Y, Wimmerova M, Sykorova J, Hynkova K, Damborsky J (2001) Biochemical characterization of broad-specificity enzymes using multivariate experimental design and a colorimetric microplate assay: characterization of the haloalkane dehalogenase mutants. J Microbiol Meth 44:149–157
Mazumdar PA, Hulecki JC, Cherney MM, Garen CR, James MNG (2008) X-ray crystal structure of Mycobacterium tuberculosis haloalkane dehalogenase Rv2579. Biochim Biophys Acta 1784:351–362
McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40(Pt 4):658–674
Morris RJ, Perrakis A, Lamzin VS (2003) ARP/wARP and automatic interpretation of protein electron density maps. Meth Enzymol 374:229–244
Nagata Y, Miyauchi K, Damborsky J, Manova K, Ansorgova A, Takagi M (1997) Purification and characterization of a haloalkane dehalogenase of a new substrate class from a gamma-hexachlorocyclohexane-degrading bacterium, Sphingomonas paucimobilis UT26. Appl Environ Microbiol 63:3707–3710
Newman J, Peat TS, Richard R, Kan L, Swanson PE, Affholter JA, Holmes IH, Schindler JF, Unkefer CJ, Terwilliger TC (1999) Haloalkane dehalogenases: structure of a Rhodococcus enzyme. Biochemistry 38:16105–16114
Oakley AJ, Klvana M, Otyepka M, Nagata Y, Wilce MC, Damborsky J (2004) Crystal structure of haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26 at 0.95 A resolution: dynamics of catalytic residues. Biochemistry 43:870–878
Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J, Sussman JL, Verschueren KHG, Goldman A (1992) The alpha/beta-hydrolase fold. Protein Eng 5:197–211
Otyepka M, Damborsky J (2002) Functionally relevant motions of haloalkane dehalogenases occur in the specificity-modulating cap domains. Protein Sci 11:1206–1217
Petrek M, Otyepka M, Banas P, Kosinova P, Koca J, Damborsky J (2006) CAVER: a new tool to explore routes from protein clefts, pockets and cavities. BMC Bioinform 7:316
Pflugrath JW (1999) The finer things in X-ray diffraction data collection. Acta Crystallogr D Biol Crystallogr 55(Pt 10):1718–1725
Pikkemaat MG, Janssen DB (2002) Generating segmental mutations in haloalkane dehalogenase: a novel part in the directed evolution toolbox. Nucleic Acids Res 30:E35
Pries F, van den Wijngaard AJ, Bos R, Pentenga M, Janssen DB (1994) The role of spontaneous cap domain mutations in haloalkane dehalogenase specificity and evolution. J Biol Chem 269:17490–17494
Prokop Z, Damborsky J, Oplustil F, Jesenska A, Nagata Y (2006a) Method of detoxification of yperite by using haloalkane dehalogenases. US Patent WO/2006/128390
Prokop Z, Oplustil F, DeFrank J, Damborsky J (2006b) Enzymes fight chemical weapons. Biotechnol J 1:1370–1380
Prokop Z, Damborsky J, Janssen DB, Nagata Y (2009) Method of production of optically active halohydrocarbons and alcohols using hydrolytic dehalogenation catalysed by haloalkane dehalogenases. US Patent WO/2006/079295
Prokop Z, Sato Y, Brezovsky J, Mozga T, Chaloupkova R, Koudelakova T, Jerabek P, Stepankova V, Natsume R, van Leeuwen JG, Janssen DB, Florian J, Nagata Y, Senda T, Damborsky J (2010) Enantioselectivity of haloalkane dehalogenases and its modulation by surface loop engineering. Angew Chem Int Ed 49:6111–6115
Schrag JD, Winkler FK, Cygler M (1992) Pancreatic lipases: evolutionary intermediates in a positional change of catalytic carboxylates? J Biol Chem 267:4300–4303
Sfetsas CC, Milios L, Skopelitou K, Venieraki A, Todou R, Flemetakis E, Katinakis P, Labrou NE (2009) Characterization of 1,2-dibromoethane-degrading haloalkane dehalogenase from Bradyrhizobium japonicum USDA110. Enzyme Microb Technol 45:397–404
Shimkets L, Ferriera S, Johnson J, Kravitz S, Beeson K, Sutton G, Rogers Y-H, Friedman R, Frazier M, Venter JC (2007) Direct Submission to NCBI database.
Stucki G, Thüer M (1995) Experiences of a large-scale application of 1,2-dichloroethane degrading microorganisms for groundwater treatment. Environ Sci Technol 29:2339–2345
Swanson PE (1999) Dehalogenases applied to industrial-scale biocatalysis. Curr Opin Biotechnol 10:365–369
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN (2004) REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr 60(Pt 12 Pt 1):2184–2195
Verschueren KHG, Franken SM, Rozeboom HJ, Kalk KH, Dijkstra BW (1993a) Refined X-ray structures of haloalkane dehalogenase at pH 6.2 and pH 8.2 and implications for the reaction-mechanism. J Mol Biol 232:856–872
Verschueren KHG, Kingma J, Rozeboom HJ, Kalk KH, Janssen DB, Dijkstra BW (1993b) Crystallographic and fluorescence studies of the interaction of haloalkane dehalogenase with halide-ions—studies with halide compounds reveal a halide binding-site in the active-site. Biochemistry 32:9031–9037
Verschueren KHG, Seljee F, Rozeboom HJ, Kalk KH, Dijkstra BW (1993c) Crystallographic analysis of the catalytic mechanism of haloalkane dehalogenase. Nature 363:693–698
Acknowledgments
We thank the German Research Foundation (DFG, Grant Bo1862/4-1) for their financial support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Martin Hesseler and Xenia Bogdanović contributed equally to this work.
Electronic supplementary materials
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hesseler, M., Bogdanović, X., Hidalgo, A. et al. Cloning, functional expression, biochemical characterization, and structural analysis of a haloalkane dehalogenase from Plesiocystis pacifica SIR-1. Appl Microbiol Biotechnol 91, 1049–1060 (2011). https://doi.org/10.1007/s00253-011-3328-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3328-x