Abstract
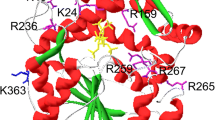
The maximum activity of Yersinia enterocolitica phytase (YeAPPA) occurs at pH 5.0 and 45 °C, and notably, its specific activity (3.28 ± 0.24 U mg−1) is 800-fold less than that of its Yersinia kristeensenii homolog (YkAPPA; 88% amino acid sequence identity). Sequence alignment and molecular modeling show that the arginine at position 79 (Arg79) in YeAPPA corresponding to Gly in YkAPPA as well as other histidine acid phosphatase (HAP) phytases is the only non-conserved residue near the catalytic site. To characterize the effects of the corresponding residue on the specific activities of HAP phytases, Escherichia coli EcAPPA, a well-characterized phytase with a known crystal structure, was selected for mutagenesis—its Gly73 was replaced with Arg, Asp, Glu, Ser, Thr, Leu, or Tyr. The results show that the specific activities of all of the corresponding EcAPPA mutants (17–2,400 U mg−1) were less than that of the wild-type phytase (3,524 U mg−1), and the activity levels were approximately proportional to the molecular volumes of the substituted residues’ side chains. Site-directed replacement of Arg79 in YeAPPA (corresponding to Gly73 of EcAPPA) with Ser, Leu, and Gly largely increased the specific activity, which further verified the key role of the residue at position 79 for determining phytase activity. Thus, a new determinant that influences the catalytic efficiency of HAP phytases has been identified.



Similar content being viewed by others
References
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modeling. Bioinformatics 22:195–201
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Fu D, Huang H, Luo H, Wang Y, Yang P, Meng K, Bai Y, Wu N, Yao B (2008) A highly pH-stable phytase from Yersinia kristeensenii: cloning, expression, and characterization. Enzyme Microb Technol 42:499–505
Fu D, Huang H, Meng K, Wang Y, Luo H, Yang P, Yuan T, Yao B (2009) Improvement of Yersinia frederiksenii phytase performance by a single amino acid substitution. Biotechnol Bioeng 103:857–864
Garrett JB, Kretz KA, O'Donoghue E, Kerovuo J, Kim W, Barton NR, Hazlewood GP, Short JM, Robertson DE, Gray KA (2004) Enhancing the thermal tolerance and gastric performance of a microbial phytase for use as a phosphate-mobilizing monogastric-feed supplement. Appl Environ Microbiol 70:3041–3046
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723
Holman WI (1943) A new technique for the determination of phosphorus by the molybdenum blue method. Biochem J 37:256–259
Huang H, Luo H, Yang P, Meng K, Wang Y, Yuan T, Bai Y, Yao B (2006) A novel phytase with preferable characteristics from Yersinia intermedia. Biochem Biophys Res Commun 350:884–889
Jenuth JP (2000) The NCBI. Publicly available tools and resources on the Web. Methods Mol Biol 132:301–312
Kemme PA, Radcliffe JS, Jongbloed AW, Mroz Z (1997) The effects of sow parity on digestibility of proximate components and minerals during lactation as influenced by diet and microbial phytase supplementation. J Anim Sci 75:2147–2153
Kim T, Mullaney EJ, Porres JM, Roneker KR, Crowe S, Rice S, Ko T, Ullah AH, Daly CB, Welch R, Lei XG (2006) Shifting the pH profile of Aspergillus niger PhyA phytase to match the stomach pH enhances its effectiveness as an animal feed additive. Appl Environ Microbiol 72:4397–4403
Kostrewa D, Gruninger-Leitch F, D'Arcy A, Broger C, Mitchell D, van Loon AP (1997) Crystal structure of phytase from Aspergillus ficuum at 2.5 Å resolution. Nat Struct Biol 4:185–190
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lehmann M, Lopez-Ulibarri R, Loch C, Viarouge C, Wyss M, van Loon AP (2000) Exchanging the active site between phytases for altering the functional properties of the enzyme. Protein Sci 9:1866–1872
Lei XG, Porres JM, Mullaney EJ, Brinch-Pedersen H (2007) Phytase: source, structure and application. In: Polaina J, MacCabe AP (eds) Industrial enzymes: source, structure and application, 1st edn. Springer, Netherlands, pp 505–529
Lim D, Golovan S, Forsberg CW, Jia Z (2000) Crystal structures of Escherichia coli phytase and its complex with phytate. Nat Struct Biol 7:108–113
Lineweaver H, Burk D (1934) The determination of enzyme dissociation constants. J Am Chem Soc 56:658–666
Li J, Li C, Xiao W, Yuan D, Wan G, Ma L (2008) Site-directed mutagenesis by combination of homologous recombination and DpnI digestion of the plasmid template in Escherichia coli. Anal Biochem 373:389–391
Luo H, Yao B, Yuan T, Wang Y, Shi X, Wu N, Fan Y (2004) Overexpression of Escherichia coli phytase with high specific activity. Chin J Biotechnol 20:78–84
Mullaney EJ, Daly CB, Ullah AH (2000) Advances in phytase research. Adv Appl Microbiol 47:157–199
Nayeem A, Chiang SJ, Liu SW, Sun Y, You L, Basch J (2009) Engineering enzymes for improved catalytic efficiency: a computational study of site mutagenesis in epothilone-B hydroxylase. Protein Eng Des Sel 22:257–266
Reddy NR, Sathe SK, Salunkhe DK (1982) Phytates in legumes and cereals. Adv Food Res 28:1–92
Selle PH, Ravindran V (2007) Microbial phytase in poultry nutrition. Anim Feed Sci Technol 135:1–41
Tomschy A, Wyss M, Kostrewa D, Vogel K, Tessier M, Hofer S, Burgin H, Kronenberger A, Remy R, van Loon AP, Pasamontes L (2000a) Active site residue 297 of Aspergillus niger phytase critically affects the catalytic properties. FEBS Lett 472:169–172
Tomschy A, Tessier M, Wyss M, Brugger R, Broger C, Schnoebelen L, van Loon AP, Pasamontes L (2000b) Optimization of the catalytic properties of Aspergillus fumigatus phytase based on the three-dimensional structure. Protein Sci 9:1304–1311
Tomschy A, Brugger R, Lehmann M, Svendsen A, Vogel K, Kostrewa D, Lassen SF, Burger D, Kronenberger A, van Loon AP, Pasamontes L, Wyss M (2002) Engineering of phytase for improved activity at low pH. Appl Environ Microbiol 68:1907–1913
Ullah AH, Gibson DM (1987) Extracellular phytase (E.C. 3.1.3.8) from Aspergillus ficuum NRRL 3135: purification and characterization. Prep Biochem 17:63–91
Ullah AH, Cummins BJ, Dischinger HC Jr (1991) Cyclohexanedione modification of arginine at the active site of Aspergillus ficuum phytase. Biochem Biophys Res Commun 178:45–53
van Etten RL, Davidson R, Stevis PE, MacArthur H, Moore DL (1991) Covalent structure, disulfide bonding, and identification of reactive surface and active site residues of human prostatic acid phosphatase. J Biol Chem 266:2313–2319
Wodzinski RJ, Ullah AH (1996) Phytase. Adv Appl Microbiol 42:263–302
Yi Z, Kornegay ET, Ravindran V, Denbow DM (1996) Improving phytate phosphorus availability in corn and soybean meal for broilers using microbial phytase and calculation of phosphorus equivalency values for phytase. Poult Sci 75:240–249
Zhang W, Lei XG (2008) Cumulative improvements of thermostability and pH-activity profile of Aspergillus niger PhyA phytase by site-directed mutagenesis. Appl Microbiol Biotechnol 77:1033–1040
Acknowledgements
This research was supported by the Key Program of Transgenic Plant Breeding (2009ZX08019-002B) and the Agricultural Science and Technology Conversion Funds (2008GB23260388) and the Earmarked Fund for Modern Agro-industry Technology Research System (NYCYTX-42-G2-05).
Author information
Authors and Affiliations
Corresponding author
Additional information
Dawei Fu and Zhongyuan Li contributed equally to this paper.
Rights and permissions
About this article
Cite this article
Fu, D., Li, Z., Huang, H. et al. Catalytic efficiency of HAP phytases is determined by a key residue in close proximity to the active site. Appl Microbiol Biotechnol 90, 1295–1302 (2011). https://doi.org/10.1007/s00253-011-3171-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3171-0