Abstract
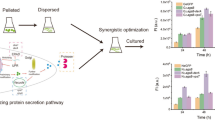
Proteolytic degradation by secreted proteases into the culture medium is one of the significant problems to be solved in heterologous protein production by filamentous fungi including Aspergillus oryzae. Double (tppA, and pepE) and quintuple (tppA, pepE, nptB, dppIV, and dppV) disruption of protease genes enhanced human lysozyme (HLY) and bovine chymosin (CHY) production by A. oryzae. In this study, we used a quintuple protease gene disruptant and performed successive rounds of disruption for five additional protease genes (alpA, pepA, AopepAa, AopepAd, and cpI), which were previously investigated by DNA microarray analyses for their expression. Gene disruption was performed by pyrG marker recycling with a highly efficient gene-targeting background (∆ligD) as previously reported. As a result, the maximum yields of recombinant CHY and HLY produced by a decuple protease gene disruptant were approximately 30% and 35%, respectively, higher than those produced by a quintuple protease gene disruptant. Thus, we successfully constructed a decuple protease gene disruptant possessing highly improved capability of heterologous protein production. This is the first report on decuple protease gene disruption that improved the levels of heterologous protein production by the filamentous fungus A. oryzae.



Similar content being viewed by others
References
Archer DB, Peberdy JF (1997) The molecular biology of secreted enzyme production by fungi. Crit Rev Biotechnol 17:273–306
Blinkovsky AM, Byun T, Brown KM, Golightly EJ (1999) Purification, characterization, and recombinant expression in Fusarium venenatum of a novel serine-type carboxypeptidase from Aspergillus oryzae. Appl Environ Microbiol 65:3298–3303
Cheevadhanarak S, Renno DV, Saunders G, Holt G (1991) Cloning and selective overexpression of an alkaline protease-encoding gene from Aspergillus oryzae. Gene 108:151–155
Foltmann B (1970) Prochymosin and chymosin (prorennin and rennin). Meth Enzymol 19:421–436
Foltmann B (1971) The biochemistry of prorennin (prochymosin) and rennin (chymosin). In: McKenzie HA (ed) The milk proteins vol. 2. Academic Press, New York, pp 217–254
Gomi K, Arikawa K, Kamiya N, Kitamoto K, Kumagai C (1993) Cloning and nucleotide sequence of the acid protease-encoding gene (pepA) from Aspergillus oryzae. Biosci Biotechnol Biochem 57:1095–1100
Gouka RJ, Punt PJ, van den Hondel CAMJJ (1997) Efficient production of secreted proteins by Aspergillus: progress, limitations and prospects. Appl Microbiol Biotechnol 47:1–11
Idiris A, Tohda H, Bi KW, Isoai A, Kumagai H, Giga-Hama Y (2006) Enhanced productivity of protease-sensitive heterologous proteins by disruption of multiple protease genes in the fission yeast Schizosaccharomyces pombe. Appl Microbiol Biotechnol 73:404–420
Jin FJ, Maruyama J, Juvvadi PR, Arioka M, Kitamoto K (2004a) Adenine auxotrophic mutants of Aspergillus oryzae: development of a novel transformation system with triple auxotrophic hosts. Biosci Biotechnol Biochem 68:656–662
Jin FJ, Maruyama J, Juvvadi PR, Arioka M, Kitamoto K (2004b) Development of a novel quadruple auxotrophic host transformation system by argB gene disruption using adeA gene and exploiting adenine auxotrophy in Aspergillus oryzae. FEMS Microbiol Lett 239:79–85
Jin FJ, Watanabe T, Juvvadi PR, Maruyama J, Arioka M, Kitamoto K (2007) Double disruption of the proteinase genes, tppA and pepE, increases the production level of human lysozyme by Aspergillus oryzae. Appl Microbiol Biotechnol 76:1059–1068
Kimura S, Maruyama J, Takeuchi M, Kitamoto K (2008) Monitoring global gene expression of proteases and improvement of human lysozyme production in the nptB gene disruptant of Aspergillus oryzae. Biosci Biotechnol Biochem 72:499–505
Kitamoto K (2002) Molecular biology of the Koji molds. Adv Appl Microbiol 51:129–153
Kitano H, Kataoka K, Furukawa K, Hara S (2002) Specific expression and temperature-dependent expression of the acid protease-encoding gene (pepA) in Aspergillus oryzae in solid-state culture (rice-koji). J Biosci Bioeng 93:563–567
Mabashi Y, Kikuma T, Maruyama J, Arioka M, Kitamoto K (2006) Development of a versatile expression plasmid construction system for Aspergillus oryzae and its application to visualization of mitochondria. Biosci Biotechnol Biochem 70:1882–1889
Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto K, Arima T, Akita O, Kashiwagi Y, Abe K, Gomi K, Horiuchi H, Kitamoto K, Kobayashi T, Takeuchi M, Denning DW, Galagan JE, Nierman WC, Yu J, Archer DB, Bennett JW, Bhatnagar D, Cleveland TE, Fedorova ND, Gotoh O, Horikawa H, Hosoyama A, Ichinomiya M, Igarashi R, Iwashita K, Juvvadi PR, Kato M, Kato Y, Kin T, Kokubun A, Maeda H, Maeyama N, Maruyama J, Nagasaki H, Nakajima T, Oda K, Okada K, Paulsen I, Sakamoto K, Sawano T, Takahashi M, Takase K, Terabayashi Y, Wortman JR, Yamada O, Yamagata Y, Anazawa H, Hata Y, Koide Y, Komori T, Koyama Y, Minetoki T, Suharnan S, Tanaka A, Isono K, Kuhara S, Ogasawara N, Kikuchi H (2005) Genome sequencing and analysis of Aspergillus oryzae. Nature 438:1157–1161
Maruyama J, Kitamoto K (2008) Multiple gene disruptions by marker recycling with highly efficient gene-targeting background (∆ligD) in Aspergillus oryzae. Biotechnol Lett 30:1811–1817
Moralejo FJ, Cardoza RE, Gutierrez S, Sisniega H, Faus I, Martin JF (2000) Overexpression and lack of degradation of thaumatin in an aspergillopepsin A-defective mutant of Aspergillus awamori containing an insertion in the pepA gene. Appl Microbiol Biotechnol 54:772–777
Morita H, Okamoto A, Yamagata Y, Kusumoto K, Koide Y, Ishida H, Takeuchi M (2009) Heterologous expression and characterization of CpI, OcpA, and novel serine-type carboxypeptidase OcpB from Aspergillus oryzae. Appl Microbiol Biotechnol 85:335–346
Murakami K, Ishida Y, Masaki A, Tatsumi H, Murakami S, Nakano E, Motai H, Kawabe H, Arimura H (1991) Isolation and characterization of the alkaline protease gene of Aspergillus oryzae. Agric Biol Chem 55:2807–2811
Nakajima K, Asakura T, Maruyama J, Morita Y, Oike H, Shimizu-Ibuka A, Misaka T, Sorimachi H, Arai S, Kitamoto K, Abe K (2006) Extracellular production of neoculin, a sweet-tasting heterodimeric protein with taste-modifying activity, by Aspergillus oryzae. Appl Environ Microbiol 72:3716–3723
Nemoto T, Watanabe T, Mizogami Y, Maruyama J, Kitamoto K (2009) Isolation of Aspergillus oryzae mutants for heterologous protein production from a double proteinase gene disruptant. Appl Microbiol Biotechnol 82:1105–1114
Punt PJ, Schuren FH, Lehmbeck J, Christensen T, Hjort C, van den Hondel CA (2008) Characterization of the Aspergillus niger prtT, a unique regulator of extracellular protease encoding genes. Fungal Genet Biol 45:1591–1599
Sharon H, Hagag S, Osherov N (2009) Transcription factor PrtT controls expression of multiple secreted proteases in the human pathogenic mold Aspergillus fumigatus. Infect Immun 77:4051–4060
van den Hombergh JP, van de Vondervoort PJ, Fraissinet-Tachet L, Visser J (1997) Aspergillus as a host for heterologous protein production: the problem of proteases. Trends Biotechnol 15:256–263
Wang Y, Xue W, Sims AH, Zhao C, Wang A, Tang G, Qin J, Wang H (2008) Isolation of four pepsin-like protease genes from Aspergillus niger and analysis of the effect of disruptions on heterologous laccase expression. Fungal Genet Biol 45:17–27
Yoon J, Kimura S, Maruyama J, Kitamoto K (2009) Construction of quintuple protease gene disruptant for heterologous protein production in Aspergillus oryzae. Appl Microbiol Biotechnol 82:691–701
Yoon J, Aishan T, Maruyama J, Kitamoto K (2010) Enhanced production and secretion of heterologous proteins by the filamentous fungus Aspergillus oryzae via disruption of vacuolar protein sorting receptor gene Aovps10. Appl Environ Microbiol 76:5718–5727
Zheng XF, Kobayashi Y, Takeuchi M (1998) Construction of a low-serine-type-carboxypeptidase-producing mutant of Aspergillus oryzae by the expression of antisense RNA and its use as a host for heterologous protein secretion. Appl Microbiol Biotechnol 49:39–44
Acknowledgments
We thank Toshiko Tanaka for experimental help. This study was supported by a Grant-in-Aid for Scientific Research (S) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and by the Program for the Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) of Japan.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary materials
Below is the link to the electronic supplementary material.
Fig. S1
a Southern blot analysis of the pepA disruptant. The open boxes (1.4 kb each) are the flanking regions for disruption of the pepA gene. The 0.3 kb upstream flanking region of the pepA ORF (hatched box) was attached at 5′-end of the downstream flanking regions, introducing direct repeats. The genomic DNAs were digested with BamHI and SacI. The strain analyzed in the panels exhibited the expected band pattern for disruption of the pepA gene. “P” and “Δ” represents the parental strain (NSPlD-tApEnBdIVdVaA1) and the gene disruptant (NSlD-tApEnBdIVdVaApA1), respectively. b Southern blot analysis of the pyrG-excised strains. The genomic DNAs were digested with BamHI and SacI. The strain analyzed in the panels exhibited the expected band pattern for excision of the pyrG gene from the pepA locus. “P” and “pyrG-” represents the parental strain (NSlD-tApEnBdIVdVaApA1) and the pyrG-excised (NSPlD-tApEnBdIVdVaApA1), respectively. c Southern blot analysis of the AopepAa disruptant. The open boxes (1.5 and 1.4 kb) are the flanking regions for disruption of the AopepAa gene. The 0.4 kb upstream flanking region of the AopepAa ORF (hatched box) was attached at 5′-end of the downstream flanking regions, introducing direct repeats. The genomic DNAs were digested with ScaI and PvuI. The strain analyzed in the panels exhibited the expected band pattern for disruption of the AopepAa gene. “P” and “Δ” represents the parental strain (NSPlD-tApEnBdIVdVaApA1) and the gene disruptant (NSlD-tApEnBdIVdVaApApAa1), respectively. d Southern blot analysis of the pyrG-excised strains. The genomic DNAs were digested with ScaI and PvuI. The strain analyzed in the panels exhibited the expected band pattern for excision of the pyrG gene from the AopepAa locus. “P” and “pyrG-” represents the parental strain (NSlD-tApEnBdIVdVaApApAa1) and the pyrG-excised (NSPlD-tApEnBdIVdVaApApAa1), respectively. e Southern blot analysis of the AopepAd disruptant. The open boxes (1.5 kb each) are the flanking regions for disruption of the AopepAd gene. The 0.3 kb upstream flanking region of the AopepAd ORF (hatched box) was attached at 5′-end of the downstream flanking regions, introducing direct repeats. The genomic DNAs were digested with EcoT22I and SacI. The strain analyzed in the panels exhibited the expected band pattern for disruption of the AopepAd gene. “P” and “Δ” represents the parental strain (NSPlD-tApEnBdIVdVaApApAa1) and the gene disruptant (NSlD-tApEnBdIVdVaApApAapAd1), respectively. f Southern blot analysis of the pyrG-excised strains. The genomic DNAs were digested with EcoT22I and SacI. The strain analyzed in the panels exhibited the expected band pattern for excision of the pyrG gene from the AopepAd locus. “P” and “pyrG-” represents the parental strain (NSlD-tApEnBdIVdVaApApAapAd1) and the pyrG-excised (NSPlD-tApEnBdIVdVaApApAapAd1), respectively. g Southern blot analysis of the cpI disruptant. The open boxes (1.5 kb each) are the flanking regions for disruption of the cpI gene. The 0.3 kb upstream flanking region of the cpI ORF (hatched box) was attached at 5′-end of the downstream flanking regions, introducing direct repeats. The genomic DNAs were digested with NdeI and ScaI. The strain analyzed in the panels exhibited the expected band pattern for disruption of the cpI gene. “P” and “Δ” represents the parental strain (NSPlD-tApEnBdIVdVaApApAapAd1) and the gene (NSlD-tApEnBdIVdVaApApAapAdcI1), respectively (PDF 222 kb)
Rights and permissions
About this article
Cite this article
Yoon, J., Maruyama, Ji. & Kitamoto, K. Disruption of ten protease genes in the filamentous fungus Aspergillus oryzae highly improves production of heterologous proteins. Appl Microbiol Biotechnol 89, 747–759 (2011). https://doi.org/10.1007/s00253-010-2937-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-010-2937-0