Abstract
In the genome of Aspergillus oryzae, 12 genes have been predicted to encode serine-type carboxypeptidases. However, the carboxypeptidase activities of the proteins encoded by these genes have not yet been confirmed experimentally. In this study, we have constructed three of these 12 genes overexpressing strains using Aspergillus nidulans and characterized their overproduced recombinant proteins. Of these three genes, one was previously named cpI; the other two have not been reported yet, and hence, we named them ocpA and ocpB. The recombinant proteins released amino acid residues from the C terminus of peptides, and the activity of the enzymes was inhibited by phenylmethylsulfonyl fluoride, indicating the enzymes to be serine-type carboxypeptidases. Recombinant OcpA, OcpB, and CpI were stable at 45°C, 55°C, and 55°C, respectively, at a low pH. The enzymatic properties of recombinant OcpB were different from those of any reported serine-type carboxypeptidase. On the other hand, recombinant OcpA had similar enzymatic properties to A. oryzae carboxypeptidases O1 and O2. The DNA and N-terminal amino acid sequences of carboxypeptidases O1 and O2 from A. oryzae IAM2640 were similar to those of OcpA. Result of transcriptional analysis of ocpA, ocpB, and cpI suggest differences in transcriptional regulation between these genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Serine-type carboxypeptidase is an exopeptidase that has Ser, His, and Asp residues as a catalytic triad construct and can sequentially release amino acid residues from the C terminus of peptides and proteins. This enzyme is widely distributed in higher plants, animals, and fungi (Doan and Fincher 1988; Dal Degan et al. 1992; Bullock et al. 1996; Shimizu et al. 1999).
Aspergillus oryzae is an important fungus in the production of Japanese traditional fermented foods, such as Japanese sake (rice wine), miso (soy bean paste), and shoyu (soy sauce). Moreover, it can produce amylases and proteases abundantly and thus is also used in the production of industrial enzymes. Recently, the genome-wide sequencing and annotation project of A. oryzae RIB40 has been completed and published (Machida et al. 2005). In this project, 12 genes have been predicted to encode serine-type carboxypeptidases because amino acid sequences deduced from those genes have serine-type carboxypeptidase-conserved motifs. However, the carboxypeptidase activities of the products of those genes have not been confirmed experimentally.
Several carboxypeptidases from A. oryzae have been purified and characterized (Nakadai et al. 1972a, b, c, 1973; Takeuchi and Ichishima 1986; Takeuchi et al. 1982; Blinkovsky et al. 1999). A. oryzae carboxypeptidase S1, which is one of the characterized serine-type carboxypeptidases from A. oryzae, is a 67,000-Da protein (Blinkovsky et al. 1999). The gene encoding this enzyme has been cloned and named cpI (National Center for Biotechnology Information (NCBI) accession no. AF394242), and its amino acid sequences has been deduced (NCBI accession no. AAK77166). Basic Local Alignment Search Tool analysis using the A. oryzae RIB40 genome data base (Database of Genomes Analyzed at NITE (DOGAN); http://www.bio.nite.go.jp/dogan/MicroTop?GENOME_ID=ao) has indicated that CpI corresponds to one of the predicted serine-type carboxypeptidases (DOGAN accession no. AO090103000026). However, genes encoding other characterized carboxypeptidases from A. oryzae have not yet been cloned. Thus, it is not clear which predicted serine-type carboxypeptidase gene of A. oryzae RIB40 encodes the characterized carboxypeptidase. The pair of gene information and encoded protein characteristics is important for the commercial application of enzymes. Therefore, the characterization of the predicted serine-type carboxypeptidase from A. oryzae RIB40 is important.
In this study, we had constructed strains overexpressing each of the predicted A. oryzae serine-type carboxypeptidase genes using Aspergillus nidulans as the host strain and found that proteins encoded by AO090103000026 (CpI), AO090012000706, and AO090701000220 have marked carboxypeptidase activities and are accumulated abundantly in liquid medium. We purified and characterized two heterologously expressed proteins encoded by AO090012000706 and AO090701000220. We also purified and characterized heterologously expressed CpI to obtain additional enzymatic properties.
Materials and methods
Strains and plasmid
A. oryzae RIB40, which was used for the genome-wide sequencing project, and A. nidulans A89 were used in this study. The plasmid vector used for constructing the overexpression plasmid was pIECS3, which had a modified A. oryzae amyB promoter upstream of the multicloning site (Fig. 1). This promoter is induced strongly by starch or maltose (Tani et al. 2000; Gomi et al. 2000) and is effective promoter in A. nidulans (Japanese, US, and European Unexamined Patent Application No. is P2003-319786A, US 2005/170453 A1, and EP1489175, respectively).
Plasmid used for construction of overexpression vectors. The modified A. oryzae amyB promoter and amyB terminator are located upstream and downstream of a multicloning site, respectively. The factor Xa recognition site and His-tag are positioned between the multicloning site and the amyB terminator. aurA r is an aureobasidin A resistance gene from A. nidulans
Media and culture conditions
A. nidulans A89 was cultured on 0.5-mM arginine-containing potato dextrose agar medium or 0.5-mM arginine-containing MYPL medium (2% maltose (w/v), 1% yeast extract (w/v), and 1% Bacto peptone (w/v); pH 5.6) at 30°C. In transformation experiments, protoplasts of the transformant candidates were regenerated in synthetic-defined (SD) selection medium (1% polypeptone S (Wako Pure chemical, Osaka, Japan; w/v), 2% glucose (w/v), 4.68% NaCl (w/v), 2% agar (w/v), 0.5 mM arginine, and 2 μg/ml Aureobasidin A (TaKaRa Bio, Shiga, Japan)) at 37°C. To obtain overexpressed proteins from transformants, the transformants were cultured in an induction medium (5% soluble starch (w/v), 1% yeast extract (w/v), 2% Bacto peptone (w/v), 0.5% KH2PO4 (w/v), 0.5% MgSO4 (w/v), 1% rice bran (w/v), and 0.5 mM arginine; pH 3.5) at 30°C. For semiquantitative reverse transcriptase polymerase chain reaction (RT-PCR) analysis, mRNA was extracted from hyphae cultured in the following media: CDGN (3% glucose (w/v), 0.3% NaNO3 (w/v), 0.1% K2HPO4 (w/v), 0.05% MgSO4 (w/v), 0.05% KCl (w/v), and 0.001% FeSO4 (w/v); pH 3.5), CDSN (CDGN medium with 3% starch (w/v) instead of 3% glucose), CDGM (CDGN medium with 0.3% skim milk (w/v) instead of 0.3% NaNO3), and CDGMN (CDGN medium with 0.3% skim milk added).
Plasmids construction
ocpA, ocpB, and cpI were amplified by PCR using the primer pairs AOS10-1F and AOS10-1R, AOS10-2F and AOS10-2R, and AOS10-3F and AOS10-3R, respectively (Table 1). The genomic DNA obtained from A. oryzae RIB40 was used as template DNA. After purifying the PCR products with the gel extract purification kit (QIAGEN, Hilden, Germany), they were digested with SacI and NotI, followed by ligation in the SacI–NotI site of the pIECS3 vector. The constructed ocpA, ocpB, and cpI expression vectors were named pIECS3ocpA, pIECS3ocpB, and pIECS3cpI, respectively.
Construction of overexpressing strains
We used A. nidulans A89 as the host strain for the construction of carboxypeptidase overexpressing strains because A. nidulans has lower proteases background levels than A. oryzae. pIECS3ocpA, pIECS3ocpB, or pIECS3cpI was introduced into A. nidulans A89 by the protoplast–polyethylene glycol method using SD selection medium. None of the expression vectors were linearized for transformation. The colony grown on SD selection medium were picked up and then confirmed by the PCR method using the primer pair up100-F and hstg2-R whether the expression plasmid was inserted into the genomic DNA (Table 1).
Carboxypeptidase activity assay
Carboxypeptidase activity was measured by the ninhydrin method described in our previous papers (Ichishima 1972; Takeuchi and Ichishima 1986). One millimolar Z-Glu-Tyr dissolved in 50 mM acetate buffer (pH 3.7) was used as substrate. The amount of Tyr liberated from Z-Glu-Tyr was determined as follows: 250 μl of the samples diluted with 50 mM acetate buffer (pH 3.7) and 250 μl of the substrate were mixed and incubated at 30°C for 20 min. After incubation, 250 μl of 0.3 M NaOH was added to terminate the reaction, and then 250 μl of 2.5% acetic acid and 1 ml of 0.5 M sodium citric acid buffer (pH 5.0) were added. Then, 500 μl of ninhydrin solution was added, and the mixture was heated at 100°C for 15 min and immediately cooled in an ice water bath. The absorbance of the mixture was measured at 570 nm. From a previous study, 1 kat of carboxypeptidase is defined as the amount of enzyme required to liberate 1 mol of Tyr from Z-Glu-Tyr per second at 30°C and pH 3.7 (Takeuchi et al. 1982). The carboxypeptidase activity for angiotensin I was investigated by matrix assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS). Two to 20 ng of the enzyme and 10 μl of 0.1 mM angiotensin I (pH 3.7) were mixed and incubated at 30°C for 3, 10, 30, 60, 120, 180, and 240 min. Three microliters of the reaction mixture was mixed with matrix solution (saturated α-cyano-4-hydroxycinnamic acid solution in 50% acetonitrile) and then applied to Voyager DE-STR and analyzed using Data Explorer software (Applied Biosystems, Foster City, CA, USA).
Purification and analysis of overexpressed proteins
Harvesting of overexpressed proteins
For recombinant protein production, the constructed ocpA-, ocpB-, and cpI-overexpressing strains were cultured in 1 or 2 l of induction medium at 30°C by rotary shaking culture for 6 days. Filtrate from the culture medium was collected, the pH of which was then adjusted to 5.5. Then, 3 g of unswelling DEAE sephadex A50 was added to the filtrate. After allowing the filtrate to stand at 4°C for 20 h, swelling DEAE sephadex A50 was collected, and then proteins were eluted with 100 ml of 50 mM sodium acetate buffer containing 500 mM sodium chloride (pH 3.5). Of ammonium sulfate, 66.2 g was added to the eluate, and the resulting precipitate was collected by centrifugation (13,000×g at 4°C for 45 min). The precipitate was dissolved in 10 mM acetate buffer (pH 3.5) and desalted using a Hitrap Desalting column (GE Healthcare, Buckinghamshire, UK). The elution step was performed with 10 mM acetate buffer (pH 3.5). The carboxypeptidase activity for Z-Glu-Tyr of each eluted fraction was assayed as described above. The active fractions from the ocpA- and ocpB-overexpressing strains were dialyzed against 10 mM sodium acetate buffer (pH 5.0) and that from the cpI-overexpressing strain was dialyzed against 10 mM sodium acetate buffer (pH 5.3).
Anion-exchange chromatography
The dialysate was applied on a Hitrap Q FF column (GE Healthcare) equilibrated with 10 mM sodium acetate buffer. The enzyme was eluted with a linear gradient of 0 to 200 mM NaCl at a flow rate of 0.5 ml/min, and 2 ml fractions were collected. Fractions containing high carboxypeptidase activities were dialyzed against 5 mM sodium acetate buffer and then pooled.
Cation-exchange chromatography
Five millimolars sodium acetate buffer (pH 3.5) was used for cation-exchange chromatography for recombinants OcpA and OcpB, and 5 mM sodium acetate buffer (pH 4.0) was used for chromatography for recombinant CpI. The pooled active fractions were applied on a Hitrap SP FF column (GE Healthcare) equilibrated with the sodium acetate buffer. The enzyme was eluted in the same manner as in anion-exchange chromatography. Active fractions were pooled and subjected to gel filtration chromatography.
Gel filtration chromatography
The active enzyme was applied on a 1.6 × 60-cm HiPrep 16/60 Sephacryl S-200 HR column (GE Healthcare) equilibrated with 10 mM sodium acetate buffer (pH 5.0). The elution was performed with the same buffer. The active fraction was pooled as purified enzyme.
Peptide mass fingerprinting
Using 10–20 μl of the purified enzyme, sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE; 10% polyacrylamide) was performed. Protein was stained with Coomassie Brilliant Blue or silver nitrate. In-gel tryptic digestion, peptide mass measurement, and analysis of fingerprints were performed as previously described (Zhu et al. 2004; Nguyen et al. 2005).
Deglycosylation study
The deglycosylation of purified recombinant proteins was performed using endoglycosidase H (Roche, Basel, Switzerland) in accordance with the manufacturer’s instructions.
Characterization of recombinant carboxypeptidases
pH optima and pH stability
The effect of pH on enzyme activity was measured in the pH range of 2.0–6.0. The pH stability of the activity was studied in the pH range of 2.0–9.0. The enzymes were incubated in different buffers having a pH range of 2.0–9.0 at 30°C for 60 min. After incubation, the enzyme activity for Z-Glu-Tyr was measured at pH 3.7 as described above.
Thermal stability
The thermal stability of the recombinant carboxypeptidases was examined in the temperature range of 30–60°C. The enzymes were incubated at each temperature in 50 mM sodium acetate buffer (pH 3.7) for 30 min, and then the residual activity for Z-Glu-Tyr at 30°C was measured as described above.
Substrate specificity
The carboxypeptidase activity of the purified recombinant enzymes for N-acyl-peptides was measured at pH 3.7 by ninhydrin assay as described above. Using Z-Gly-Lys, Bz-Gly-Arg, Z-Phe-Tyr-Leu, or Z-Phe-Leu as substrate, the activity was assayed at pH 5.0. For kinetic analysis, purified recombinant carboxypeptidases were tested with Z-Glu-Tyr and Z-Leu-Tyr as substrates at various concentrations at 30°C in 50 mM acetate buffer (pH 3.7). K m and k cat were calculated using the Lineweaver–Burk plot. Protein concentration was measured spectrophotometrically at 280 nm. The molar extinction coefficients were calculated using the equation described previously (Pace et al. 1995)
Inhibition study
The effects of protease inhibitors on recombinant carboxypeptidase activity were studied using phenylmethylsulfonyl fluoride (PMSF), N-[N-(l-3-trans-carboxirane-2-carbonyl)-l-leucyl]-agmatine (E64), pepstatin A, ethylenediaminetetraacetic acid (EDTA), and monoiodoacetic acid (MIA) at final concentrations of 1, 0.1 mM, 3 μg/ml, 1, and 1 mM, respectively. The mixture of the enzyme with each inhibitor was incubated at pH 3.7 and 30°C for 10 min, and then residual activity for Z-Glu-Tyr was measured.
Protein N-terminal sequencing and DNA sequencing
For the N-terminal amino acid sequencing, purified carboxypeptidases O1 and O2 produced by A. oryzae IAM2640 (Takeuchi et al 1982) were used as samples. The samples were separated by SDS-PAGE. After electrophoresis, the separated proteins were transferred to polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA), and then their N-termini were sequenced by Edman degradation on an ABI 476A protein sequencer (Applied Biosystems, Foster City, CA, USA). For the nucleotide sequencing of ocpA homolog from A. oryzae IAM2640, ocpA homolog was amplified by PCR using the primer pair AOS10-1F and AOS10-1R (Table 1). The genomic DNA obtained from A. oryzae IAM2640 was used as template DNA. The amplified sample was digested with SacI and NotI and then ligated in the SacI–NotI site of the pBluescript II vector. The constructed plasmid was used as template DNA for DNA cycle sequencing with primers specific for ocpA: ocpAseqF1, ocpAseqF2, ocpAseqF3, ocpAseqF4, ocpAseqF5, ocpAseqF6, ocpAseqR1, ocpAseqR2, ocpAseqR3, and ocpAseqR4 (Table 1). DNA cycle sequencing was performed using the DYEnamic ET Terminator Cycle Sequencing kit (GE Healthcare) on an ABI 310 Genetic Analyzer (Applied Biosystems).
Semiquantitative RT-PCR analysis
mRNA was purified from total RNA using Oligotex-dT30 super (TaKaRa Bio, Shiga, Japan) in accordance with the manufacturer’s instructions. The primer pairs AOS10-1F and AOS10-1R, AOS10-2F and AOS10-2R, and AOS10-3F and AOS10-3R were used to amplify ocpA, ocpB, and cpI, respectively, and the primer pair actin-F and actin-R was used to amplify the gene encoding γ-actin (Table 1). PCR was performed for 26 or 32 cycles using a step-cycle program of 96°C for 30 s, 60°C for 30 s, and 72°C for 3 min.
Results
Construction of predicted carboxypeptidase overexpressing strains
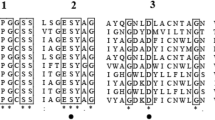
The deduced amino acid sequence of the protein encoded by the genes AO090012000706, AO090701000220, and AO090103000026 (cpI) has the conserved substrate binding motif and active site motifs of serine-type carboxypeptidases (Fig. 2). AO090012000706 and AO090701000220 were named ocpA (A. oryzae carboxypeptidase) and ocpB, respectively. OcpA showed a high amino acid sequence homology with Penicillium janthinellum carboxypeptidase S1 (identity, 69%), whereas OcpB showed a low amino acid sequence homology with any of the reported carboxypeptidases. Overall, OcpA exhibited 35% and 37% amino acid identities to OcpB and CpI, respectively. The amino acid identity between OcpB and CpI was 42%.
Multiple alignment of the amino acid sequences of AO090012000706, AO090701000220, AO090103000026 (CpI), and three known serine-type carboxypeptidases: S. cerevisiae carboxypeptidase Y (CPY; NP_014026), Aspergillus phoenicis CpdS (P52719), and P. janthinellum carboxypeptidase S1 (CPD-S1; AAB28596). The sequence of CPD-S1 contains no preprosequence. The conserved substrate binding motif is indicated by the line (I), and catalytic motifs harboring the amino acid of the catalytic triad (filled lozenge) are marked II–IV. S1 binding sites are shown by filled reverse triangles, S1′ binding sites by filled circles, and completely conserved residues by asterisks
To determine their enzymatic properties, OcpA, OcpB, and CpI were overproduced in A. nidulans. ocpA-, ocpB-, and cpI-overexpressing strains were constructed by transformation using pIECS3ocpA, pIECS3ocpB, and pIECS3cpI, respectively (see “Materials and methods”). The resulting strains, which were termed A89CS3ocpA, A89CS3ocpB, and A89CS3cpI, were inoculated into the induction medium. After 6 days of culture, SDS-PAGE showed that the media from the culture of the transformants contained a specific protein abundantly (Fig. 3a) and that they had a significant protease activity for Z-Glu-Tyr, whereas that from the culture of the host strain showed scarcely detectable activity. Because we found another putative start codon at the 134th nucleotide upstream from the start codon of ocpB, we also constructed a re-anotated ocpB-overexpressing strain. However, the media from the culture of the transformant showed no carboxypeptidase activity. Therefore, we used the strains A89CS3ocpA, A89CS3ocpB, and A89CS3cpI for the enzyme production.
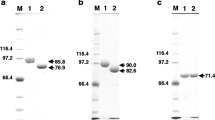
SDS-PAGE profile using culture media (a) and purified recombinant proteins from transformants as samples (b). The proteins contained in culture media and the purified proteins were separated using a 10% SDS gel and visualized with silver nitrate or Coomassie Brilliant Blue. a Media from the culture of A89CS3ocpA (lane 1), A89CS3ocpB (lane 2), A89CS3cpI (lane 3), and A. nidulans A89 (lane 4). bArrowed bands were excised from the gel for identification by peptide mass fingerprinting. Bands appeared at 30 kDa in lanes 3, 5, and 7 correspond to endoglycosidase H. Lane 1 Molecular weight marker; lanes 2, 4, and 6 purified recombinants OcpA, OcpB, and CpI, respectively; lanes 3, 5, and 7 deglycosylated purified recombinants OcpA, OcpB, and CpI, respectively. Deglycosylated recombinant OcpA was shown as two bands (filled arrowheads)
Purification of recombinant carboxypeptidases
Since the overexpression vector pIECS3 had a His-tag coding region behind a multicloning site, we determined whether a His-tag was added at the C terminus of overproduced proteins by Western blot analysis using the media from the culture of the transformants as samples. The result showed that the overproduced proteins had no His-tag. Therefore, the overproduced proteins from the transformants were purified on anion-exchange, cation-exchange, and gel filtration columns. After cultivating A89CS3ocpA, A89CS3ocpB, and A89CS3cpI at 30°C for 6 days, the filtrate from the culture media showed carboxypeptidase activities of 10.5, 4.4, and 23.7 nkat/ml, respectively. Purification led to the acquisition of 0.56 and 0.46 mg of purified protein with specific activities of 355.2 and 57.6 nkat/mg from 670 and 710 ml of media from the cultures of A89CS3ocpA and A89CS3ocpB, respectively. From 1,440 ml of media from the culture of A89CS3cpI, 0.41 mg of purified protein with 275.0 nkat/mg was obtained.
The purified proteins, which were indicated by arrows A, B, and C in Fig. 3b, were identified by peptide mass fingerprinting using MALDI-TOF MS and the MASCOT search program. The result indicated that the proteins were the target proteins, i.e., the proteins indicated by arrows A, B, and C were the gene products of ocpA, ocpB, and cpI, respectively. Deglycosylated recombinant OcpA was shown as two bands, consisting of one band corresponding to a protein of 52,000 Da and one band corresponding to a protein with a slightly lower molecular mass (Fig. 3b, lane 3). The peptide mass fingerprinting showed that both bands corresponded to proteins encoded by ocpA.
As shown in Fig. 3b, SDS-PAGE showed that purified recombinants OcpA, OcpB, and CpI had no contaminants; their apparent molecular masses, as calculated from the SDS-PAGE results, were 58,000, 126,000, 90,000 Da, respectively (Fig. 3b). Deglycosylated purified recombinants OcpA, OcpB, and CpI were consistent with their molecular masses calculated from their deduced amino acid sequences (i.e., 52,614, 67,480, and 61,141 Da, respectively).
Enzymatic properties of recombinants OcpA, OcpB, and CpI
When purified recombinants OcpA, OcpB, and CpI were incubated with angiotensin I (Asp-Arg-Val-Tyr-Ile-His-Pro-Phe-His-Leu) at 30°C, mass peaks derived from the degradation products Asp-Arg-Val-Tyr-Ile-His-Pro-Phe-His and Asp-Arg-Val-Tyr-Ile-His-Pro-Phe were detected by MALDI-TOF-MS (Fig. 4). Similarly, when bradykinin (Arg-Pro-Pro-Gly-Phe-Ser-Pro-Phe-Arg) was used as substrate, a mass peak derived from the product from which a C-terminal amino acid was released (Arg-Pro-Pro-Gly-Phe-Ser-Pro-Phe) was detected (data not shown). Using angiotensin I and bradykinin as substrates, recombinants OcpA, OcpB, and CpI released an amino acid residue sequentially from the C terminus of the substrates, indicating that these recombinant proteins were carboxypeptidases.
MS scan analysis of carboxypeptidase activities of recombinants OcpA, OcpB, and CpI. a MS spectrum of angiotensin I. MS spectra of b recombinant-OcpA-digested peptide, c recombinant-OcpB-digested peptide, and d recombinant-CpI-digested peptide. Of recombinants OcpA, OcpB, and CpI, 2, 20, and 17 ng, respectively, were mixed with 1 nmol of angiotensin I, and then the mixture was incubated at 30°C for 180, 10, and 60 min, respectively. The peak at m/z 1,299 correspond to whole angiotensin I (arrows). The two peaks at m/z 1,184 and 1,047 correspond to Asp-Arg-Val-Tyr-Ile-His-Pro-Phe-His (gray arrowheads) and Asp-Arg-Val-Tyr-Ile-His-Pro-Phe (black arrowheads), respectively
The effect of pH on enzyme activity was determined at 30°C using Z-Glu-Tyr as substrate (Table 2). The optimal pH of recombinants OcpA and CpI was pH 4.0 and that of OcpB was within pH 3.5 to 3.7. The pH stability of these enzymes was examined in the pH range of 2–9. Recombinants OcpA, OcpB, and CpI showed residual activities of over 60% relative to their maximum activities in the pH ranges 4–7, 3–8, and 3–8, respectively. Recombinant CpI maintained activities of 40% and 50% of the maximum activity after incubation at pHs 2 and 9, respectively, whereas recombinants OcpA and OcpB scarcely showed activity after incubation at such pHs.
The thermal stabilities of above recombinant carboxypeptidases were studied between 30°C and 60°C. Residual activity was measured after exposure at each temperature for 30 min (Table 2). The activity of recombinant OcpA scarcely decreased at 40°C; however, at 50°C, the activity decreased to less than 1% of the maximum. On the other hand, the decline in the activities of recombinants OcpB and CpI was only 6% after exposure at 50°C. The activities of these two enzymes decreased to less than 10% after exposure at 60°C.
The substrate specificities of the purified recombinant enzymes was examined with small N-acylpeptides as substrate (Table 3), and the kinetics parameters for Z-Glu-Tyr and Z-Leu-Tyr are shown in Table 4. Recombinant OcpA showed high activities for Z-Glu-Tyr, Z- Phe-Leu, and Z-Phe-Tyr-Leu, while recombinant OcpB exhibited strong activities for Z-Leu-Tyr, Z-Tyr-Leu, Z-Phe-Tyr-Leu, and Z-Gly-Pro-Leu-Gly. The best substrate for recombinants OcpA and CpI was Z-Phe-Leu and that for recombinant OcpB was Z-Phe-Tyr-Leu. Recombinant OcpA did not prefer peptides containing Gly at the P1 position, and recombinant OcpB was able to hydrolyze substrates that had a basic amino acid residue at the P1′ position. The kinetic parameters for Z-Glu-Tyr and Z-Lue-Tyr showed that recombinant OcpB had a weak hydrolysis ability compared with recombinants OcpA and CpI. The k cat/K m value of recombinant OcpA was higher for Z-Glu-Tyr than for Z-Leu-Tyr; in contrast, those of recombinants OcpB and CpI were lower for Z-Glu-Tyr than for Z-Leu-Tyr. Recombinant OcpA had similar substrate specificities to carboxypeptidases O1 and O2, which have been reported by Takeuchi et al (1982).
The effects of protease inhibitors on protease activity were examined using PMSF, E64, MIA, pepstatin A, and EDTA. The serine protease inhibitor PMSF inhibited the activities of all the recombinant enzymes (Fig. 5). MIA inhibited the activities of recombinants OcpA and CpI, as well as of carboxypeptidases O1 and O2, but not the activity of recombinant OcpB. Pepstatin A and EDTA did not affect the activity of recombinant OcpA, OcpB, or CpI.
Recombinant OcpA exhibited similar enzymatic properties to carboxypeptidases O1 and O2, particularly in terms of substrate specificity and the activity inhibition rate with an inhibitor.
Effects of protease inhibitors on the carboxypeptidase activities of recombinants OcpA, OcpB, and CpI. PMSF, E64, MIA, pepstatin A, and EDTA were added to the reaction mixture at final concentrations of 1, 0.1 mM, 3 μg/ml, 1, and 1 mM, respectively. The activities for Z-Glu-Tyr of the enzymes with each inhibitor were measured at pH 3.7 at 30°C. Reaction mixtures containing water or dimethyl sulfoxide instead of each inhibitor were used as reference. Relative activities were quantified as the percentage activity of each reference. Open bar recombinant OcpA; filled bar recombinant OcpB; hatched bar recombinant CpI
N-terminal sequence analysis of carboxypeptidases O1 and O2 and DNA sequence analysis of ocpA homolog from A. oryzae IAM2640
To investigate the similarity of carboxypeptidases O1 and O2 and OcpA, the N-terminal amino acid sequences of these enzymes and the nucleotide sequence of the ocpA homolog from A. oryzae IAM2640 were determined and compared with those of OcpA and ocpA from A. oryzae RIB40. As shown in Fig. 6, the N-terminal sequences of carboxypeptidases O1 and O2 were completely identical, that is, YVXNSGIXET. This sequence showed a high identity with the sequence of the N-terminal region of OcpA deduced from the nucleotide sequence of ocpA from A. oryzae RIB40 (Fig. 6). In addition, the entire nucleotide sequence, including predicted introns, of ocpA homolog from A. oryzae IAM2640 was the same as that of ocpA from A. oryzae RIB40.
Amino acid sequences of N-terminal regions of carboxypeptidases O1 and O2 and OcpA. The N-terminal sequence of carboxypeptidases O1 and O2 were determined using an amino acid sequencer. That of OcpA was deduced from the nucleic acid sequence of ocpA from A. oryzae RIB40 (DOGAN accession no. AO090012000706). Asterisks show completely conserved amino acid residues
Transcriptional analysis of ocpA, ocpB, and cpI from A. oryzae wild-type strain
To investigate what nutrient condition induces the transcription of ocpA, ocpB, and cpI in the A. oryzae wild-type strain, the transcription levels of these genes were examined by semiquantitative RT-PCR analysis. For this analysis, mRNA was isolated from mycelia of A. oryzae RIB40 cultured in liquid media with glucose or soluble starch as the sole carbon source and NaNO3 and/or skim milk as the nitrogen source. As shown in Fig. 7, ocpB was transcribed at equal levels in all media. On the other hand, the transcriptions of ocpA and cpI were dependent on the nitrogen source. The mRNA transcription level of ocpA was relatively low when the strain was harvested from the media with NaNO3 as the sole nitrogen source (CDSN and CDGN media; Fig. 7, lanes 5 and 6); however, it greatly increased when the strain was harvested from the media with skim milk (CDGM and CDGMN media; Fig. 7, lanes 7 and 8). The transcription level of cpI modestly increased in the CDGM medium (Fig. 7, lane 15); however, it was not affected in the NaNO3–skim milk-containing medium unlike that of ocpA (CDGMN medium; Fig. 7, lane 16). The transcriptional upregulation of ocpA and cpI was also detected in the medium with gelatin as the sole nitrogen source (data not shown).
Semiquantitative RT-PCR analysis of ocpA, ocpB, and cpI in A. oryzae RIB40. mRNA was isolated from approximately 0.1 g (semidry wet) of mycelia cultured for 1 day in CDSN (lanes 1, 5, 9, 13), CDGN (lanes 2, 6, 10, 14), CDGM (lanes 3, 7, 11, 15), or CDGMN (lanes 4, 8, 12, 16) media. Each medium contained different carbon and nitrogen sources. CDSN contained 3% starch and 0.3% NaNO3, CDGN contained 3% glucose and 0.3% NaNO3, CDGM contained 3% glucose and 0.3% skim milk, and CDGMN contained 3% glucose, 0.3%NaNO3, and 0.3% skim milk. PCR for amplification of ocpA, ocpB, and cpI was performed using 32 cycles and that for γ-actin was performed using 26 cycles. The PCR amplification of γ-actin using each cDNA as a template was carried out for quantitative control. Negative-control PCR analysis using each mRNA as a template showed no amplified fragments (data not shown)
Discussion
In this study, we carried out the heterologous expression and purification of CpI and two predicted serine-type carboxypeptidases from A. oryzae RIB40 and experimentally determined that the proteins have carboxypeptidase activity.
To facilitate the purification of the overexpressed proteins, the plasmid vector which had a His-tag coding region behind a multicloning site was used for construction of ocpA-, ocpB-, and cpI-overexpressing strains. However, the overproduced proteins secreted in the medium had no His-tag, which might have been cleaved at the factor Xa recognition site by the endopeptidase from the host strain during posttranslational processes.
Purified recombinants OcpA, OcpB, and CpI released amino acid residues sequentially from the C-termini of angiotensin I and bradykinin. Moreover, the protease activities of these recombinant enzymes were inhibited by the serine protease inhibitor PMSF. These results indicate that these proteins are serine-type carboxypeptidases.
Several carboxypeptidases from A. oryzae have been reported (Nakadai et al. 1972a, b, c, 1973; Takeuchi and Ichishima 1986; Takeuchi et al. 1982; Blinkovsky et al. 1999). Carboxypeptidases O1 and O2 produced by A. oryzae IAM2640 have been reported to have similar enzymatic properties except for their activities for angiotensin (Takeuchi et al. 1982). Recombinant OcpA is identical or similar in enzymatic properties to carboxypeptidases O1 and O2: (a) These enzymes have strong hydrolysis activities for Z-Glu-Tyr, Z-Tyr-Leu, and Z-Gly-Pro-Leu-Gly, but the activities for Z-Gly-Lys, Z-Gly-Phe, and Z-Gly-Leu are negligible levels. (b) The protease activities of the enzymes are completely inhibited by MIA. (c) The molecular weights of the enzymes not subjected to endoglycosidase H treatment are approximately 60,000 Da. In addition, the amino acid sequences of the N-terminal regions of OcpA and carboxypeptidases O1 and O2 are almost completely identical, and the ocpA homolog from A. oryzae IAM2640 exhibits 100% identity to ocpA from A. oryzae RIB40. Moreover, SDS-PAGE and peptide mass finger printing analysis showed that the translational product of ocpA had two mature forms with different molecular weights. These results indicate that the translational products of ocpA mature into two enzymes through C-terminal processing and that such mature enzymes are equivalent to carboxypeptidases O1 and O2.
The enzymatic properties of recombinant OcpB differed from those of any previously reported carboxypeptidase from A. oryzae. Homologs of ocpB are present in the genome of other fungi, including A. nidulans, Aspergillus fumigatus, Aspergillus clavatus, and Aspergillus niger; however, proteins encoded by these homologs have not yet been characterized. Thus, this is the first report on the enzymatic properties of the gene product of ocpB. The comparison of the amino acid sequence of OcpA, OcpB, and CpI with that of carboxypeptidase Y, which is one of the well-characterized serine-type carboxypeptidases (Endrizzi et al. 1994; Jung et al. 1995, 1999), led to the prediction that amino acid residues localize in the subsites S1 and S1′ of OcpA, OcpB, and CpI (Fig. 2). These amino acid residues of OcpA and OcpB are very similar, with a difference of only one amino acid residue that form subsite S1 and that is located at the second posterior position from the active Asp residue. In OcpA, an Ile residue is located at this position; on the other hand, in OcpB is an Ala residue. Since Ala has a shorter side chain than Ile, the conformation of subsite S1 will differ between OcpA and OcpB, and this spatial difference may be one of the causes of substrate specificity.
Carboxypeptidase Y has a Cys residue (Cys341) around its catalytic Asp residue. Cys341 forms an S1 subsite and is not essential for enzyme activity; however, it affects on catalytic efficiency by controlling the size of the S1 substrate binding pocket (Jung et al. 1999). The result of our inhibition study showed that the hydrolysis of Z-Glu-Tyr by recombinants OcpA and CpI was completely inhibited by the cystein protease inhibitor MIA. OcpA, OcpB, and CpI have a Cys residue corresponding to the Cys341 of carboxypeptidase Y around the predicted catalytic Asp residue (Fig. 2). One possible reason for the inhibition of the activity of serine-type carboxypeptidase by cystein protease inhibitors is the alkylation of this Cys residue. The reduction in hydrolysis ability may be caused by the blockade of the interaction between the enzyme and the substrate, resulting from the alkylation of the Cys residue at the subsite by a cystein protease inhibitor. Unlike those of recombinants OcpA and CpI, the activity of recombinant OcpB was scarcely inhibited by MIA. In addition, the effect of PMSF on the activity of recombinant OcpB was small compared with those on the activities of recombinants OcpA and CpI. These results suggest that the three-dimensional structure of OcpB is different from those of OcpA and CpI; consequently, inhibitors will not be able to enter the substrate binding pocket.
Protein glycosylation has been shown to be related to protein stability (Wujek et al. 2004; Solá and Griebenow 2008; Yasuda et al. 1999; Jafari-Aghdam et al. 2005). The apparent molecular masses of recombinants OcpB and CpI, as calculated from mobility data on SDS-PAGE profiles, were approximately 126,000 and 90,000 Da, whereas the molecular masses calculated from the deduced amino acid sequences were 67,480 and 61,141 Da, respectively. These results indicate that carbohydrate molecular masses of recombinants OcpB and CpI are approximately 46% and 32% of the total molecular masses of these enzymes, respectively. On the other hand, that of recombinant OcpA is only 9% of the total molecular mass. The low percentage of glycosylation may be one of the factors for the instability of this enzyme.
The transcription level of ocpA from the A. oryzae wild-type strain markedly increased in the growth medium with an exogenous protein. The upregulation of this gene also occurred when both sodium nitrate and protein were present in the medium, indicating that the transcriptional regulation of this gene is dependent on presence of an exogenous protein but not on the absence of the preferred nitrogen source. The mRNA transcription level of cpI modestly increased in the presence of the exogenous protein; however, unlike ocpA, the mRNA level of cpI did not increase in the medium containing both sodium nitrate and protein. The regulation of ocpB was not dependent on the presence of an exogenous protein. These results suggest that the regulatory pathways for the transcription of the three genes are different.
Carboxypeptidases can sequentially release amino acids from the C terminus of peptides and proteins; hence, they are useful tools for C terminus sequencing in the protein studies and debittering and flavor-improving processes in the food industry. Recombinants OcpA, OcpB, and CpI have different substrate specificities, and theses enzymes accumulate in liquid medium abundantly. These properties are useful for the application to commerce and industrial production of these enzymes.
References
Blinkovsky AM, Byun T, Brown KM, Golightly EJ (1999) Purification, characterization, and recombinant expression in Fusarium venenatum of a novel serine-type carboxypeptidase from Aspergillus oryzae. Appl Environ Microbiol 65:3298–3303
Bullock TL, Breddam K, Remington SJ (1996) Peptide aldehyde complexes with wheat serine-type carboxypeptidase II: implications for the catalytic mechanism and substrate specificity. J Mol Biol 255:714–725
Dal Degan F, Ribadeau-Dumas B, Breddam K (1992) Purification and characterization of two serine-type carboxypeptidases from Aspergillus niger and their use in C-terminal sequencing of proteins and peptide synthesis. Appl Environ Microbiol 58:2144–2152
Doan NP, Fincher GB (1988) The A- and B-chains of carboxypeptidase I from germinated barley originate from a single precursor polypeptide. J Biol Chem 263:11106–11110
Endrizzi JA, Breddam K, Remington SJ (1994) 2.8-A structure of yeast serine carboxypeptidase. Biochem 33:11106–11120
Gomi K, Akeno T, Minetoki T, Ozeki K, Kumagai C, Okazaki N, Iimura Y (2000) Molecular cloning and characterization of a transcriptional activator gene, amyR, involved in the amylolytic gene expression in Aspergillus oryzae. Biosci Biotechnol Biochem 64:816–827
Ichishima E (1972) Purification and characterization of a new type of acid carboxypeptidase from Aspergillus. Biochim Biophys Acta 258:274–288
Jafari-Aghdam J, Khajeh K, Ranjbar B, Nemat-Gorgani M (2005) Deglycosylation of glucoamylase from Aspergillus niger: effects on structure, activity and stability. Biochim Biophys Acta 1750:61–68
Jung G, Ueno H, Hayashi R, Liao TH (1995) Identification of the catalytic histidine residue participating in the charge-relay system of carboxypeptidase Y. Protein Sci 4:2433–2435
Jung G, Ueno H, Hayashi R (1999) Carboxypeptidase Y: structural basis for protein sorting and catalytic triad. J Biochem 126:1–6
Machida M, Asai K, Sano M, Tanaka T, Kumagai T et al (2005) Genome sequencing and analysis of Aspergillus oryzae. Nature 438:1157–1161
Nakadai T, Nasuno S, Iguchi N (1972a) Purification and properties of acid carboxypeptidase I from Aspergillus oryzae. Agr Biol Chem 36:1343–1352
Nakadai T, Nasuno S, Iguchi N (1972b) Purification and properties of acid carboxypeptidase II from Aspergillus oryzae. Agr Biol Chem 36:1473–1480
Nakadai T, Nasuno S, Iguchi N (1972c) Purification and properties of acid carboxypeptidase III from Aspergillus oryzae. Agr Biol Chem 36:1481–1488
Nakadai T, Nasuno S, Iguchi N (1973) Purification and properties of acid carboxypeptidase IV from Aspergillus oryzae. Agr Biol Chem 37:1237–1251
Nguyen CH, Tsurumizu R, Sato T, Takeuchi M (2005) Taka-amylase A in the conidia of Aspergillus oryzae RIB40. Biosci Biotechnol Biochem 69:2035–2041
Pace CN, Vajdos F, Fee L, Grimsley G, Gray T (1995) How to measure and predict the molar absorption coefficient of a protein. Protein Sci 4:2411–2423
Shimizu H, Ueno H, Hayashi R (1999) Role of carbohydrate moiety in carboxypeptidase Y: structural study of mutant enzyme lacking carbohydrate moiety. Biosci Biotechnol Biochem 63:1045–1050
Solá RJ, Griebenow K (2008) Effects of glycosylation on the stability of protein pharmaceuticals. J Pharm Sci 98(4):1223–1245
Takeuchi M, Ushijima T, Ichishima E (1982) A new acid carboxypeptidase, O-1, from Aspergillus oryzae. Curr Microbiol 7:19–23
Takeuchi M, Ichishima E (1986) A 155K acid carboxypeptidase O from Aspergillus oryzae. Agri Biol Chem 50:633–638
Tani S, Kawaguchi T, Kato M, Kobayashi T, Tsukagoshi N (2000) A novel nuclear factor, SREB, binds to a cis-acting element, SRE, required for inducible expression of the Aspergillus oryzae Taka-amylase A gene in A. nidulans. Mol Gen Genet 263:232–238
Zhu LY, Nguyen CH, Sato T, Takeuchi M (2004) Analysis of secreted proteins during conidial germination of Aspergillus oryzae RIB40. Biosci Biotechnol Biochem 68:2607–2612
Wujek P, Kida E, Walus M, Wisniewski KE, Golabek AA (2004) N-glycosylation is crucial for folding, trafficking, and stability of human tripeptidyl-peptidase I. J Biol Chem 279:12827–12839
Yasuda Y, Ikeda S, Sakai H, Tsukuba T, Okamoto K, Nishishita K, Akamine A, Kato Y, Yamamoto K (1999) Role of N-glycosylation in cathepsin E. A comparative study of cathepsin E with distinct N-linked oligosaccharides and its nonglycosylated mutant. Eur J Biochem 266:383–391
Acknowledgment
This work was supported by the Program for the Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Morita, H., Okamoto, A., Yamagata, Y. et al. Heterologous expression and characterization of CpI, OcpA, and novel serine-type carboxypeptidase OcpB from Aspergillus oryzae . Appl Microbiol Biotechnol 85, 335–346 (2009). https://doi.org/10.1007/s00253-009-2087-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-2087-4