Abstract
We investigated the diversity, spatial distribution, and abundances of ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) in sediment samples of different depths collected from a transect with different distances to mangrove forest in the territories of Hong Kong. Both the archaeal and bacterial amoA genes (encoding ammonia monooxygenase subunit A) from all samples supported distinct phylogenetic groups, indicating the presences of niche-specific AOA and AOB in mangrove sediments. The higher AOB abundances than AOA in mangrove sediments, especially in the vicinity of the mangrove trees, might indicate the more important role of AOB on nitrification. The spatial distribution showed that AOA had higher diversity and abundance in the surface layer sediments near the mangrove trees (0 and 10 m) but lower away from the mangrove trees (1,000 m), and communities of AOA could be clustered into surface and bottom sediment layer groups. In contrast, AOB showed a reverse distributed pattern, and its communities were grouped by the distances between sites and mangrove trees, indicating mangrove trees might have different influences on AOA and AOB community structures. Furthermore, the strong correlations among archaeal and bacterial amoA gene abundances and their ratio with NH +4 , salinity, and pH of sediments indicated that these environmental factors have strong influences on AOA and AOB distributions in mangrove sediments. In addition, AOA diversity and abundances were significantly correlated with hzo gene abundances, which encodes the key enzyme for transformation of hydrazine into N2 in anaerobic ammonium-oxidizing (anammox) bacteria, indicating AOA and anammox bacteria may interact with each other or they are influenced by the same controlling factors, such as NH +4 . The results provide a better understanding on using mangrove wetlands as biological treatment systems for removal of nutrients.
Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Avoid common mistakes on your manuscript.
Introduction
Mangrove ecosystems, the important natural wetlands distributed along estuaries in tropical and subtropical region, provide breeding, growing, refuge, and feeding zone for many marine organisms (Holguin et al. 2001). In mangrove ecosystem, microbial activities are the key processes contributing to the high productivity of this ecosystem (Holguin et al. 2001). Microbial-mediated nutrient transformation and subsequent export to other marine ecosystems, such as nitrogen and phosphate, are important to mangrove and other coastal ecosystems (Sjoling et al. 2005). For the nitrogen (N) cycle within mangrove ecosystem, microbial processes include dinitrogen (N2)-fixation, nitrification, denitrification, ammonification, anaerobic ammonium oxidizing (anammox), and dissimilatory nitrate reduction to ammonium (Purvaja et al. 2008). The alternating aerobic and anaerobic conditions caused by tidal flushing in mangrove wetlands provide a suitable environment for the nitrification (Kristensen et al. 1998) and denitrification (Rivera-Monroy and Twilley 1996) or anammox (Meyer et al. 2005), which affects the rate of N turnover. On the other hand, the mangrove trees could also enhance microbial N transformation by transporting O2 to the otherwise anoxic subsurface sediment through their aerial roots to support nitrification (Holguin et al. 2001) or providing the carbon source to fuel heterotrophic denitrification in the rhizosphere (Zhu and Sikora 1995); at the same time, mangrove trees may inhibit microbial N transformation due to the competition for available N in mangrove ecosystem. These information show that a complex microbial N transformation exists in a mangrove ecosystem which requires more research efforts to understand the dynamics of theses microorganisms and their contributions to the N cycle in mangrove ecosystem.
Nitrification, a two-step process that includes the oxidation of ammonium via hydroxylamine to nitrite and then nitrate, is a key process in marine N cycling. The first and rate-limiting step, ammonia oxidation, is executed by limited number of microbial groups, including aerobic chemoautotrophic bacteria and archaea. Bacterial members include the β-proteobacteria Nitrosomonas and Nitrosopiras, and the γ-proteobacteria Nitrosococcus (Purkhold et al. 2000). Both, the amoA genes which encode the catalytic α-subunit of the ammonia monooxygenase enzyme and 16S rRNA genes have been used in molecular studies for analyzing ammonia-oxidizing bacteria (AOB) in the environment, specifically to determine the effects on diversity, abundance and community structures by physicochemical parameters, e.g., pH, temperature, oxygen, light, soil management regimes, etc. (Kowalchuk and Stephen 2001; Prosser and Embley 2002). The first ammonia-oxidizing archaeon (AOA) Nitrosopumilus maritimus has been isolated from a marine aquarium tank recently (Konneke et al. 2005). N. maritimus is a representative of the ubiquitous marine “group 1” archaeota and grows chemolithoautotrophically by oxidizing ammonia to nitrite under mesophilic conditions (Konneke et al. 2005). In addition, N. maritimus contains putative genes for all three subunits (amoA, amoB, and amoC) of the ammonia monooxygenase (Konneke et al. 2005). The discovery of ammonia oxidizers in the archaea has inspired many researchers to investigate this new group. Francis et al. (2005) provided the first molecular evidence demonstrating the archaeal amoA gene to be pervasive in the ocean, including the euphotic zone, suboxic water columns and coastal/estuarine sediments. Up to now, the diversity, distribution and physiology of AOA have been widely investigated in soils (Adair and Schwartz 2008; Hansel et al. 2008), seawater column (Francis et al. 2005; Lam et al. 2007; Mincer et al. 2007; Beman et al. 2008), marine sediments (Beman and Francis 2006; Santoro et al. 2008), aquaria filters (Urakawa et al. 2008), rhizosphere (Chen et al. 2008; Herrmann et al. 2008, 2009), municipal sewage plants (Park et al. 2006), hot spring (de la Torre et al. 2008; Hatzenpichler et al. 2008; Zhang et al. 2008), hydrothermal vents, and even marine invertebrates (Steger et al. 2008). Erguder et al. (2009) summarized the current knowledge on the environmental conditions related to the presence of AOA and identified site-related properties and environmental factors which included ammonium, organic carbon, temperature, salinity, oxygen, pH, sulfide, phosphate, and some metal contents. Furthermore, the interactions between nitrifiers (AOA and AOB) and anammox bacteria were also recorded in the Black Sea (Lam et al. 2007) and in the Peruvian oxygen minimum zone (Lam et al. 2009), where nitrifiers were proposed to supply the nitrite to anammox to form a nitrification-anammox coupling.
It is apparent that previous studies have demonstrated convincingly that AOA and AOB are widely distributed and regulated by the physiochemical characteristics of environments, but no study has examined AOA and AOB in mangrove ecosystems. Nitrification rates in mangrove sediments measured by chemical methods indicated that nitrification occurred predominantly close to the sediment surface in micro-oxic zones, which were created by the oxygen-pumping activity of mangrove roots and in the oxidized lining of animal burrows (Holguin et al. 2001; Purvaja et al. 2008). The studies suggested nitrification rates increased generally with depth at the vegetated site, where nitrifiers were subjected to more oxic conditions compared with non-vegetated sediments, indicating the distribution of mangrove plants might have strong influences on nitrification in mangrove ecosystems (Holguin et al. 2001; Purvaja et al. 2008). Constructed and natural mangrove wetlands have been employed as alternative low cost biological systems for treatment of municipal sewage with high inorganic nitrogen, which might be transformed by AOA and AOB (Wong et al. 1997). However, the community structures and distribution of AOA and AOB, and their relationship with mangrove trees are still largely unknown. In the present study, we report the phylogenetic diversity, distribution and abundance of AOA and AOB based on archaeal and bacterial amoA genes analyses in surface and lower layer sediments collected from different sites located at different distances to mangrove trees in the New Territories of Hong Kong. In addition, the relationships of the diversity, distribution and abundance of AOA and AOB with environmental factors, mangrove trees and occurrence of copies of the hzo gene which in anammox bacteria encodes the key enzyme for transformation of hydrazine to N2 and is used as an indicator of anammox activity and potential (Strous et al. 2006; Shimamura et al. 2007; Schmid et al. 2008), were also analyzed using statistical analysis. The results will help us to improve our understanding of the usage of mangrove wetlands as wastewater treatment systems for removal of nutrients.
Materials and methods
Sample collection and physicochemical analyses
Mai Po Marshes Nature Reserve, located at the northwestern corner of the New Territories of Hong Kong (22°30′N, 114°02′E), is the largest remaining coastal wetland in Hong Kong. Mai Po Marshes Nature Reserve comprises of subtropical mangroves, inter-tidal mudflats, as well as man-made fishponds and drainage channels. In the mangrove wetland, the dominated mangrove trees are Kandelia obovata (formerly Kandelia candel). The area is quite homogenous and three sampling sites (S0, S1, and S2) were selected in a transect: site S0 was immediately to the mangrove trees in the forest; site S1 with a distance of 10 m to the mangrove forest edge, and site S2 on the inter-tidal mudflats without any mangrove tree or vegetation about 1,000 m from S0 (Electronic supplementary Fig. 1). The surface layer (1–2 cm) and lower layer (20–21 cm) sediment samples were collected in triplicate at each of the three sampling sites and homogenized the triplicate sediment samples into one to represent each site, and all samples were immediately transferred into a 4°C cooler for transport to the laboratory for analyses (within 2 h). Samples were labeled by the sampling sites (S0-S2) with surface layer (s) or lower layer (l), and obtained sequences in each site were labeled by their site name with clone number.
Temperature, redox potential and pH of the sediment samples were measured in situ using IQ180G Bluetooth Multi-Parameter System (Hach Company, Loveland, CO) and concentrations of NH +4 -N, NO −3 -N, and NO −2 -N in pore water of sediment samples were measured, after centrifugation, with an autoanalyzer (QuickChem, Milwaukee, WI) according to standard methods given by the American Public Health Association (American Public Health Association 1995). Salinity of pore water was measured using a YSI 556 Multiprobe System (YSI, Yellow Springs, OH). The results are summarized in the Electronic supplemenatry Table S-1.
DNA extraction, PCR amplification, and cloning
Total genomic DNA of each sediment samples were extracted using the SoilMaster DNA Extraction kit (Epicentre Biotechnologies, Madison, WI), respectively. Archaeal amoA genes were amplified using primers Arch-amoAF (5′-STAATGGTCTGGCTTAGACG-3′) and Arch-amoAR (5′-GCGGCCATCCATCTGTATGT-3′) (Francis et al. 2005), while bacterial amoA genes were amplified by amoA-1F (5′-GGGGTTTCTACTGGTGGT-3′) and amoA-2R (5′-CCCCTCKGSAAAGCCTTCTTC-3′) (Rotthauwe et al. 1997). Polymerase chain reaction (PCR) reaction mixtures contained 1 μl DNA (30–50 ng μl−1), 1 μl bovine serum albumin (100 mg ml−1, Roche), 5 μl 10 × GoTaq Flexi buffer (Promega, Hong Kong), and 4 μl MgCl2 (25 mM, Promega), 1 μl of dNTPs (5 mM, Invitrogen, Hong Kong), 1 μl of each forward and reverse primer (20 μM), and 0.25 μl of GoTaq Flexi polymerase (5 U μl−1, Promega, Hong Kong), and the final volume for each reaction was 50 μl. PCR reactions were performed as follows: 95°C for 3 min; 35 cycles of 95°C for 45 s, 53°C (for AOA) or 55°C (for AOB) for 1 min, followed by 72°C for 1 min; and finally 72°C for 7 min. Triplicate PCR reaction products for each site were pooled; gel purified using Gel Advance-Gel Extraction System (VIOGEME, Taipei) and cloned using the pMD18 T-vector (Takara, Japan), and replicated clone libraries were constructed for each sampling sites for the two layers. The insertion of an appropriate-sized DNA fragment was determined by PCR amplification with the primer set M13F and M13R (Invitrogen, Hong Kong).
Sequencing and phylogenetic analysis
Sequencing was performed with the Big Dye Terminate kit (Applied Sciences, Foster City, CA) and an ABI Prism 3730 DNA analyzer (Applied Biosystems, Foster City, CA). DNA sequences were examined and edited by MEGA 4.0 software (Tamura et al. 2007). In order to obtain reliable phylogenetic results, we carried out the alignment by using identical number of bp for each of the obtained sequences (465 bp for archaeal amoA and 640 bp for bacterial amoA), which were almost the full fragments amplified by the selected primer sets used in our study. Nucleic acid sequences of archaeal and bacterial amoA genes were aligned using the MEGA 4.0 software package, and phylogenetic trees were constructed with the neighbor-joining and maximum parsimony methods, and bootstrap re-sampling analysis for 1,000 replicates was performed to estimate the confidence of the tree topologies (Tamura et al. 2007).
Quantitative PCR assay
The copy numbers of amoA gene of AOA and AOB in all sediment samples were determined in triplicate using an ABI 7000 Sequence detection system (Applied Biosystems, Foster City, CA). The quantification was based on the fluorescent dye SYBR-Green I. Each reaction was performed in a 25 μl volume containing 1 μl of DNA template, 1 μl BSA (100 mg ml−1, Roche), 1 μl of each primer (20 μM) and 12.5 μl of Power SYBR-Green PCR Master Mix (Applied Biosystems, Foster City, CA). The PCR protocols were identical as those described above except that the final extension step was omitted and that 50 cycles were performed followed by melting analyses. The hzo gene abundances of anammox bacteria in the same sediment samples were also analyzed by quantitative PCR, and details for the PCR amplification experiments are available elsewhere (Li et al. 2010). We used the constructed plasmids carrying the targeted gene sequences (amoA or hzo) as the standards, which were generated by amplifying genes from extracted DNA of site S1-s sediments and cloning them into pMD18 T-vector (Takara, Japan). The abundances of targeted genes in each sample were calculated by the parallel quantitative PCR of different dilutions of the standard plasmids, where copy number of standard plasmids in each dilution could be calculated by their concentrations. The quantitative PCR amplification efficiencies ranged from 0.92–0.96 (AOA amoA), 0.91–0.95 (AOB amoA) and 0.85–0.92 (hzo), and correlation coefficients (R 2) were greater than 0.99 for the three targeted genes. Comparison for amoA gene abundances of AOA and AOB was done by analysis of variance (ANOVA) for definition of any significant differences among these sediment sample sites with their different layers.
Statistical analysis
To obtain views on the richness and diversity of archaeal and bacterial amoA genes, operational taxonomic units (OTUs) for community analysis were defined by 5% differences in nucleotide sequences, as determined by using the furthest neighbor algorithm in DOTUR (Schloss and Handelsman 2005). Shannon and Simpson indices for each sampling site were also generated by DOTUR. The community structures of archaeal and bacterial amoA genes between any two clone libraries were compared with LIBSHUFF software version 0.96 (http://libshuff.mib.uga.edu/) to get a quantitative documentation according to the Singleton method (Singleton et al. 2001). To further rigorously examine the geographic distribution of phylogenetic structures of AOA and AOB in mangrove sediments, the phylogenetic trees derived from the alignment used in Figs. 2 and 3 but restricted to archaeal and bacterial amoA sequences recovered in this study were analyzed by using the principal coordinates analysis (PCoA) and Jackknife Environment Clusters analysis in UniFrac (Lozupone et al. 2006). Finally, correlation analysis of AOA and AOB community structures with environmental factors, mangrove trees and anammox bacteria was conducted using Microsoft Excel program with Pearson’s moment correlation.
Nucleotide sequence accession numbers
The GenBank accession numbers for the archaeal and bacterial amoA genes sequences reported here are GQ331390-GQ331635 and GQ331636-GQ331908, respectively. The hzo gene accession numbers are GQ331363 to GQ331389.
Results
Richness and phylogenetic diversity of AOA and AOB
In this study, 246 archaeal and 273 bacterial amoA clones were sequenced from 12 AOA and AOB clone libraries (Table 1). Within each individual sampling site, two to eight AOA and two to eight AOB OTUs were found. In some cases, such as AOA in S1-s and AOB in S2-s, the OTU numbers would be higher if more clones were sequenced, based on non-asymptotic rarefaction curves (Electronic supplementary Fig. S-2). The highest AOA richness occurred at S1-s, and the lowest occurred in S0-l, while the highest AOB richness occurred at S2-l, and the lowest was S0-s, consisting with the values of Shannon and Simpson indices. In addition, AOA showed a higher richness at the surface layer in the vicinity of the mangrove tree (S0 and S1); while AOB had higher OTUs at site S2 than S0 or S1 for both surface and lower layers sediments (Table 1).
Phylogenetic diversity analysis showed that archaeal amoA sequences were divided into five clusters (Fig. 1). Cluster 1 fell into a phylogenetic group comprising a large proportion of archaeal amoA sequences (150 out of 246) and some published sequences recovered from deep sea, bay, or estuary sediments (Dang et al. 2008, 2009). Sequences in clusters 2 and 3 recovered from the surface layer of the research sites were more closely related to those from wastewater treatment plants (Park et al. 2006; Zhang et al. 2009). It was interesting that the sequences in cluster 4 formed a distinct phylogenetic cluster where DNA sequence identity with available amoA sequences was <88%, indicating new site specific AOA species in mangrove sediments. Approximately 10% of archaeal amoA clones fell into cluster 5 which related to amoA sequences from Northern California coastal/estuarine sediment (Mosier and Francis 2008) and N. maritimus (Konneke et al. 2005). Comparing with AOA, the phylogeny of AOB was much simpler and all amoA sequences related to Nitrosomonas-like sequences were grouped in three different clusters (Fig. 2). About 41.8% of sequences in cluster 1 were closely related to the clones recovered from Bahía del Tóbari estuaries, Chesapeake Bay and Jiaozhou Bay (Francis et al. 2003; Beman and Francis 2006). Similar to AOA, a large proportion (148 out of 273) of bacterial amoA sequences also formed a site specific group (cluster 3), and the DNA identity with available sequences was less than 89%. Furthermore, three sub-clusters could be identified within cluster 3, where sequences in two of the three sub-clusters were recovered only from site S2 and the third group was retrieved from all sampling sites.
Phylogenetic tree of archaeal amoA genes from surface (solid) and lower layers (open) mangrove sediments collected from the locations at immediately to the mangrove trees (S0), 10 m (S1), and 1,000 m (S2) away from mangrove trees. Bootstrap values (>50%) are indicated at branch points, with distance bootstrap values at front and parsimony values at back. Branch lengths correspond to sequence differences as indicated by the scale bar
Phylogenetic tree of bacterial amoA genes from surface (solid) and lower layers (open) mangrove sediments collected from the locations at immediately to the mangrove trees (S0), 10 m (S1), and 1,000 m (S2) away from mangrove trees. Bootstrap values (>50%) are indicated at branch points, with distance bootstrap values at front and parsimony values at back. Branch lengths correspond to sequence differences as indicated by the scale bar
amoA and hzo gene abundances in mangrove sediments
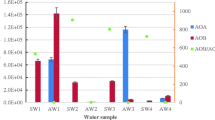
AOA amoA gene copy numbers ranged from 6.04 × 106 to 9.45 × 106 copies per dry gram of sediment at surface layers, and the S0 and S1 located near mangrove trees had higher archaeal amoA gene abundances than S2 located far away from mangrove trees. In contrast, AOA amoA gene abundances in lower layer sediments were increasing from S0 to S2 along with the distances of sampling sites to mangrove trees (Fig. 3a). However, AOB amoA gene abundances were decreasing from S0 to S2 at both surface and lower layer sediments, ranging from 0.95 × 107 to 4.17 × 107 copies per dry gram of sediment (Fig. 3b). In addition, AOA amoA gene abundances were higher at surface layer sediments for sites S0 and S1 but lower at lower layer sediments for the site S2, while AOB amoA gene abundances showed the opposite (p < 0.05, n = 3, ANOVA). The ratios of bacterial to archaeal amoA genes (0.87 ± 0.27 to 11.93 ± 0.25) indicated that AOB amoA copy numbers were greater than those of AOA in mangrove sediments except in the lower layer of S2, where was far away from mangrove trees (Fig. 3c). Furthermore, the significant different (p < 0.05, n = 3, ANOVA) ratios of AOB/AOA amoA abundance decreased from 5.01 ± 0.55 to 2.22 ± 0.69 at surface layers and 11.93 ± 0.25 to 0.86 ± 0.27 at lower layers from sites S0 to S2, respectively, which indicates a striking spatial shift of AOB/AOA amoA abundances in mangrove sediments, especially in the lower layer sediments. Except no hzo genes were detected at site S0-l, the obtained anammox hzo gene abundances ranged from 7.5 × 104 to 36.5 × 104 copies per gram of sediments (dry weight) (Fig. 3d). The hzo gene determined at sites S1 and S0 showed a significant higher copy number in surface layer samples than in lower layer samples (p < 0.05, n = 3). However, in site S2, the surface layer sample had a lower hzo gene copy number than the lower layer sample (p < 0.05, n = 3).
Community distribution of AOA and AOB in mangrove sediments
Based on the p values calculated by LIBSHUFF software, AOA showed similar community structure compositions only in samples collected at sites S0-s with S1-s or in samples collected at sites S0-s with S2-s (95% confidence), while they were significantly different for any other two clone libraries as judged from pairwise comparison, indicating an obvious shift of AOA community structure in mangrove sediments (Table 2). However, for AOB community structures, there was a significant compositional overlap among the six bacterial amoA clone libraries (95% confidence). Sites S0 and S1 for both surface and lower layers also showed similar bacterial amoA compositions which further supported a uniform AOB community structure at this research area (Table 2). However, PCoA (data not shown) and Jackknife Environment Clusters separated the AOA communities into slightly distinct surface and lower layer groups while AOB communities were separated into groups by distances of being near and far away from mangrove trees groups (Fig. 4).
Correlations of ammonia-oxidizer community structures with environmental factors and anammox bacteria communities
Pearson’s moment correlation analysis was used to find out whether there were some environmental factors affecting the distributions of AOA and AOB in mangrove sediments (Table 3). Results showed that the diversity and abundance of archaeal amoA sequences were significantly correlated with the NH +4 while presence of bacterial amoA was strongly correlated with the distances between sampling sites to mangrove trees and salinity. In addition, abundances of archaeal and bacterial amoA genes and their ratios were also significantly correlated with sediment pH. Furthermore, it was found that AOA diversity and abundance were strongly and positively correlated with the copy number of anammox bacteria hzo genes (Table 3).
Discussion
In the present study, we draw a comprehensive picture about the diversity, distribution and abundances of AOA and AOB in mangrove sediments. Diversity estimations indicated that both archaeal and bacterial richness (0.53–1.60 and 0.66–1.70 predicted by Shannon index, and 0.24–0.74 and 0.20–0.52 predicted by Simpson index) were within the ranges reported at other estuaries (0.44–2.12 and 0.33–2.02 for Shannon index; 0.10–0.84 and 0.11–0.84 for Simpson index) (Beman and Francis 2006; Mosier and Francis 2008; Santoro et al. 2008) (Table 1). However, phylogenetic analysis showed that both the distributions of archaeal and of bacterial amoA sequences supported that distinct phylogenetic groups in mangrove sediment and sequences in the distinct groups are less than 88% and 89% identified to other available sequences, which provides new information about AOA diversity and distribution in natural ecosystem (Figs. 1 and 2). However, the presence of the AOA sequences related to sequences found previously in samples taken from wastewater treatment plants of Hong Kong and the presence of AOB amoA sequences related to sequences found in Hong Kong marine aquaculture indicated that AOA and AOB in mangrove sediments of Mai Po Nature Reserve might also originate from the export of municipal wastes of the nearby area since large quantities of wastewater from domestic and industrial origins (including aquaculture) are discharged each year into Mai Po Nature Reserve without proper treatment (Wang et al. 2006).
From the abundances of AOA and AOB in mangrove sediments (Fig. 3), we found that AOA abundances in Mai Po Nature Reserve were similar to those in the San Francisco Bay estuary (Mosier and Francis 2008), while AOB abundances were slightly higher than those in salt marsh and coastal aquifer sediments (104∼106 copies per gram sediment) (Dollhopf et al. 2005; Santoro et al. 2008). However, in contrast to numerous recent studies suggesting that AOA were more abundant than AOB in marine and terrestrial ecosystems (Leininger et al. 2006; Wuchter et al. 2006; Lam et al. 2007; Mincer et al. 2007; Beman et al. 2008), AOB amoA copy numbers were greater than that of AOA amoA in mangrove sediments except in the lower layer of S2 (p < 0.05, ANOVA). Furthermore, AOB abundances were more abundant in the vicinity of the mangrove trees, where AOB abundances decreased from S0 to S2, while AOA abundances increased at the same transect (except S2-s, p < 0.05, ANOVA). These results might have two indications. One is that AOB might play a more important role in the nitrification in mangrove sediments than AOA and the other is that mangrove trees might have strong interactions with AOA and AOB, which strongly affected their distributions in mangrove sediments.
It has been reported that ammonia oxidizers often exhibit strong spatial structure distribution in estuaries. Studies in the Bahía del Tóbari and San Francisco Bay estuaries have shown that AOA communities differ between the interior and the mouth of the bays (Beman and Francis 2006; Mosier and Francis 2008). AOB communities shifted along salinity gradients in the Chesapeake Bay, Plum Island Sound and Ythan estuaries (Francis et al. 2003; Bernhard et al. 2005; Freitag et al. 2006). In the present study, AOA communities had little compositional overlap in surface layer sediments (S0-s vs S1-s and S0-s vs S2-s) but were significantly different in lower layers and their compositions could be divided into slightly distinct groups based on sediments collected from surface or lower layers (Fig. 4a). However, communities of AOB overlapped significantly at both surface and lower layer sediment samples, and were clearly separated into two groups according the distances of sites location from mangrove trees (Fig. 4b). Together the abundance variations of AOA and AOB in mangrove sediments and the distinct spatial patterns for AOA and AOB communities in mangrove sediments further confirm that the existence of mangrove trees might have strong influences on the distribution of AOA and AOB in both diversity and abundances. In previous researches, it has been shown that the presence of some plants, such as the freshwater macrophyte Littorella uniflora (Herrmann et al. 2008), rice (Chen et al. 2008), Spartina alterniflora, or Spartina patens (Moin et al. 2009) has strong influences on the diversity and abundance of AOA and AOB. Furthermore, the plants-nutrient-microorganisms interaction model in mangrove ecosystems has been presented in a previous study (Holguin et al. 2001); here, the strong influences of mangrove trees on AOA and AOB community structures and the significant correlations of the AOA diversity and abundance with the distance of sampling sites to mangrove trees might also give strong evidence on this model (Table 3).
Interactions between nitrifiers (AOA and AOB) and anammox bacteria had been recorded in the Black Sea (Lam et al. 2007) and in the Peruvian oxygen minimum zone (Lam et al. 2009), where nitrifiers were proposed to supply the nitrite to anammox to form a nitrification-anammox coupling. The significantly and positively correlations between AOA diversity and abundance with the anammox bacteria hzo gene abundances might also indicate that the complex interactions not only exist among microorganisms and mangrove forest but also within different N-transforming microorganisms. However, no similar relationships were found in between AOB and anammox bacteria, which indicated that AOB communities might be also affected by other environmental factors, such as salinity. Indeed, AOB diversity and abundances were found to be significantly correlated with salinity. Since salinity is a particularly important parameter for ammonia oxidation in coastal and estuary sediments, in part because of its influence on NH +4 adsorption in sediments (Boatman and Murray 1982; Boynton and Kemp 1985), it is thus reasonable that salinity has a strong influence on AOB which might be more important on nitrification in mangrove sediments. In addition, a positive correlation between AOA abundance and pH, and negative correlations between AOB abundance and AOB amoA/AOA amoA ratios with pH were found in the present study, indicating that sediment pH is also an important factor affecting the abundances of AOA and AOB in mangrove sediments (Table 3).
In conclusion, unique phylogenetic groups of archaeal and bacterial amoA genes were revealed in mangrove sediments, indicating site specific AOA and AOB occur in Mai Po mangrove sediments. The spatial distribution of AOA and AOB communities and abundances were strongly influenced by the presence of mangrove trees, nitrogen substrates, and anammox bacteria. Furthermore, the higher abundance of AOB than AOA indicated that AOB may play a more important role on nitrification in the mangrove sediment. The results provide more specific information on more effective use of mangrove wetlands as alternative biological systems for treatment of wastewater or surface run-off containing high inorganic nitrogen.
References
Adair KL, Schwartz E (2008) Evidence that ammonia-oxidizing archaea are more abundant than ammonia-oxidizing bacteria in semiarid soils of northern Arizona, USA. Microb Ecol 56:420–426
American Public Health Association (1995) Standard methods for the examination of water and wastewater. APHA, Washington
Beman JM, Francis CA (2006) Diversity of ammonia-oxidizing archaea and bacteria in the sediments of a hypernutrified subtropical estuary: Bahia del Tobari, Mexico. Appl Environ Microbiol 72:7767–7777
Beman JM, Popp BN, Francis CA (2008) Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J 2:429–441
Bernhard AE, Donn T, Giblin AE, Stahl DA (2005) Loss of diversity of ammonia-oxidizing bacteria correlates with increasing salinity in an estuary system. Environ Microbiol 7:1289–1297
Boatman CD, Murray JW (1982) Modeling exchangeable NH +4 adsorption in marine-sediments—process and controls of adsorption. Limnol Oceanogr 27:99–110
Boynton WR, Kemp WM (1985) Nutrient regeneration and oxygen-consumption by sediments along an estuarine salinity gradient. Mar Ecol-Prog Ser 23:45–55
Chen XP, Zhu YG, Xia Y, Shen JP, He JZ (2008) Ammonia-oxidizing archaea: important players in paddy rhizosphere soil? Environ Microbiol 10:1978–1987
Dang H, Zhang X, Sun J, Li T, Zhang Z, Yang G (2008) Diversity and spatial distribution of sediment ammonia-oxidizing crenarchaeota in response to estuarine and environmental gradients in the Changjiang Estuary and East China Sea. Microbiol 154:2084–2095
Dang H, Li J, Zhang X, Li T, Tian F, Jin W (2009) Diversity and spatial distribution of amoA-encoding archaea in the deep-sea sediments of the tropical West Pacific Continental Margin. J Appl Microbiol 106:1482–1493
de la Torre JR, Walker CB, Ingalls AE, Konneke M, Stahl DA (2008) Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ Microbiol 10:810–818
Dollhopf SL, Hyun JH, Smith AC, Adams HJ, O’Brien S, Kostka JE (2005) Quantification of ammonia-oxidizing bacteria and factors controlling nitrification in salt marsh sediments. Appl Environ Microbiol 71:240–246
Erguder TH, Boon N, Wittebolle L, Marzorati M, Verstraete W (2009) Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiol Rev 33:855–869
Francis CA, O’Mullan GD, Ward BB (2003) Diversity of ammonia monooxygenase (amoA) genes across environmental gradients in Chesapeake Bay sediments. Geobiology 1:129–140
Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB (2005) Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci U S A 102:14683–14688
Freitag TE, Chang L, Prosser JI (2006) Changes in the community structure and activity of betaproteobacterial ammonia-oxidizing sediment bacteria along a freshwater-marine gradient. Environ Microbiol 8:684–696
Hansel CM, Fendorf S, Jardine PM, Francis CA (2008) Changes in bacterial and archaeal community structure and functional diversity along a geochemically variable soil profile. Appl Environ Microbiol 74:1620–1633
Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, Wagner M (2008) A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci U S A 105:2134–2139
Herrmann M, Saunders AM, Schramm A (2008) Archaea dominate the ammonia-oxidizing community in the rhizosphere of the freshwater macrophyte Littorella uniflora. Appl Environ Microbiol 74:3279–3283
Herrmann M, Saunders AM, Schramm A (2009) Effect of lake trophic status and rooted macrophytes on community composition and abundance of ammonia-oxidizing prokaryotes in freshwater sediments. Appl Environ Microbiol 75:3127–3136
Holguin G, Vazquez P, Bashan Y (2001) The role of sediment microorganisms in the productivity, conservation, and rehabilitation of mangrove ecosystems: an overview. Biol Fertil Soils 33:265–278
Konneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA (2005) Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546
Kowalchuk GA, Stephen JR (2001) Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu Rev Microbiol 55:485–529
Kristensen E, Jensen MH, Banta GT, Hansen K, Holmer M, King GM (1998) Transformation and transport of inorganic nitrogen in sediments of a southeast Asian mangrove forest. Aquat Microb Ecol 15:165–175
Lam P, Jensen MM, Lavik G, McGinnis DF, Müller B, Schubert CJ, Amann R, Thamdrup B, Kuypers MM (2007) Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proc Natl Acad Sci U S A 104:7104–7109
Lam P, Lavik G, Jensen MM, van de Vossenberg J, Schmid M, Woebken D, Gutierrez D, Amann R, Jetten MS, Kuypers MM (2009) Revising the nitrogen cycle in the Peruvian oxygen minimum zone. Proc Natl Acad Sci U S A 106:4752–4557
Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C (2006) Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809
Li M, Hong Y, Klotz MG, Gu J-D (2010) A comparison of primer sets for detecting 16S rRNA and hydrazine oxidoreductase genes of anaerobic ammonium-oxidizing bacteria in marine sediments. Appl Microbiol Biotechnol 86:781–790
Lozupone C, Hamady M, Knight R (2006) UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371
Meyer RL, Risgaard-Petersen N, Allen DE (2005) Correlation between anammox activity and microscale distribution of nitrite in a subtropical mangrove sediment. Appl Environ Microbiol 71:6142–6149
Mincer TJ, Church MJ, Taylor LT, Preston C, Karl DM, DeLong EF (2007) Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ Microbiol 9:1162–1175
Moin NS, Nelson KA, Bush A, Bernhard AE (2009) Distribution and diversity of archaeal and bacterial ammonia oxidizers in salt marsh sediments. Appl Environ Microbiol 75:7461–7468
Mosier AC, Francis CA (2008) Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ Microbiol 10:3002–3016
Park HD, Wells GF, Bae H, Criddle CS, Francis CA (2006) Occurrence of ammonia-oxidizing archaea in wastewater treatment plant bioreactors. Appl Environ Microbiol 72:5643–5647
Prosser JI, Embley TM (2002) Cultivation-based and molecular approaches to characterisation of terrestrial and aquatic nitrifiers. Antonie Van Leeuwenhoek 81:165–179
Purkhold U, Pommerening-Roser A, Juretschko S, Schmid MC, Koops HP, Wagner M (2000) Phylogeny of all recognized species of ammonia oxidizers based on comparative 16 S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl Environ Microbiol 66:5368–5382
Purvaja R, Ramesh R, Ray AK, Rixen T (2008) Nitrogen cycling: A review of the processes, transformations and fluxes in coastal ecosystems. Curr Sci 94:1419–1438
Rivera-Monroy VH, Twilley RR (1996) The relative role of denitrification and immobilization in the fate of inorganic nitrogen in mangrove sediments (Terminos Lagoon, Mexico). Limnol Oceanogr 41:284–296
Rotthauwe JH, Witzel KP, Liesack W (1997) The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712
Santoro AE, Francis CA, de Sieyes NR, Boehm AB (2008) Shifts in the relative abundance of ammonia-oxidizing bacteria and archaea across physicochemical gradients in a subterranean estuary. Environ Microbiol 10:1068–1079
Schloss PD, Handelsman J (2005) Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71:1501–1506
Schmid MC, Hooper AB, Klotz MG, Woebken D, Lam P, Kuypers MM, Pommerening-Roeser A, Op den Camp HJ, Jetten MS (2008) Environmental detection of octahaem cytochrome c hydroxylamine/hydrazine oxidoreductase genes of aerobic and anaerobic ammonium-oxidizing bacteria. Environ Microbiol 10:3140–3149
Shimamura M, Nishiyama T, Shigetomo H, Toyomoto T, Kawahara Y, Furukawa K, Fujii T (2007) Isolation of a multiheme protein with features of a hydrazine-oxidizing enzyme from an anaerobic ammonium-oxidizing enrichment culture. Appl Environ Microbiol 73:1065–1072
Singleton DR, Furlong MA, Rathbun SL, Whitman WB (2001) Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl Environ Microbiol 67:4374–4376
Sjoling S, Mohammed SM, Lyimo TJ, Kyaruzi JJ (2005) Benthic bacterial diversity and nutrient processes in mangroves: impact of deforestation. Estuar Coast Shelf Sci 63:397–406
Steger D, Ettinger-Epstein P, Whalan S, Hentschel U, de Nys R, Wagner M, Taylor MW (2008) Diversity and mode of transmission of ammonia-oxidizing archaea in marine sponges. Environ Microbiol 10:1087–1094
Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, Taylor MW, Horn M, Daims H, Bartol-Mavel D, Wincker P, Barbe V, Fonknechten N, Vallenet D, Segurens B, Schenowitz-Truong C, Medigue C, Collingro A, Snel B, Dutilh BE, Op den Camp HJ, van der Drift C, Cirpus I, van de Pas-Schoonen KT, Harhangi HR, van Niftrik L, Schmid M, Keltjens J, van de Vossenberg J, Kartal B, Meier H, Frishman D, Huynen MA, Mewes HW, Weissenbach J, Jetten MS, Wagner M, Le Paslier D (2006) Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440:790–794
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Urakawa H, Tajima Y, Numata Y, Tsuneda S (2008) Low temperature decreases the phylogenetic diversity of ammonia-oxidizing archaea and bacteria in aquarium biofiltration systems. Appl Environ Microbiol 74:894–900
Wang Y, Leung PC, Qian PY, Gu J-D (2006) Antibiotic resistance and plasmid profile of environmental isolates of Vibrio species from Mai Po Nature Reserve, Hong Kong. Ecotoxicol 15:371–378
Wong YS, Tam NFY, Lan CY (1997) Mangrove wetlands as wastewater treatment facility: a field trial. Hydrobiologia 352:49–59
Wuchter C, Abbas B, Coolen MJ, Herfort L, van Bleijswijk J, Timmers P, Strous M, Teira E, Herndl GJ, Middelburg JJ, Schouten S, Sinninghe Damste JS (2006) Archaeal nitrification in the ocean. Proc Natl Acad Sci U S A 103:12317–12322
Zhang CL, Ye Q, Huang Z, Li W, Chen J, Song Z, Zhao W, Bagwell C, Inskeep WP, Ross C, Gao L, Wiegel J, Romanek CS, Shock EL, Hedlund BP (2008) Global occurrence of archaeal amoA genes in terrestrial hot springs. Appl Environ Microbiol 74:6417–6426
Zhang T, Jin T, Yan Q, Shao M, Wells G, Criddle C, PF HH (2009) Occurrence of ammonia-oxidizing archaea in activated sludges of a laboratory scale reactor and two wastewater treatment plants. J Appl Microbiol 107:970–977
Zhu T, Sikora FJ (1995) Ammonium and nitrate removal in vegetated and unvegetated gravel bed microcosm wetlands. Water Sci Technol 32:219–228
Acknowledgements
This research was supported in part by grants from Agriculture, Fisheries, and Conservation Department of the Hong Kong SAR Government, and Faculty of Science on Sustainable Water at The University of Hong Kong (to J-DG); National Natural Science Foundation of China (3080032), Knowledge Innovation Key Project of The Chinese Academy of Sciences (KZCX2-YW-QN207), Guangdong Province Natural Science Foundation (84510301001692), and a start-up fund for Excellent Scholarship of the Chinese Academy of Sciences (07YQ091001) (to Y-GH). We would like to thank Jessie Lai for laboratory support at The University of Hong Kong.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 370 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Li, M., Cao, H., Hong, Y. et al. Spatial distribution and abundances of ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) in mangrove sediments. Appl Microbiol Biotechnol 89, 1243–1254 (2011). https://doi.org/10.1007/s00253-010-2929-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-010-2929-0