Abstract
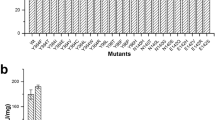
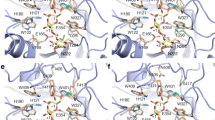
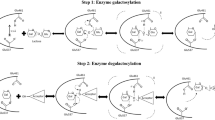
β-galactosidase is an enzyme administered as a digestive supplement to treat lactose intolerance, a genetic condition prevalent in most world regions. The gene encoding an acid-stable β-galactosidase potentially suited for use as a digestive supplement was cloned from Aspergillus niger van Tiegh, sequenced and expressed in Pichia pastoris. The purified recombinant protein exhibited kinetic properties similar to those of the native enzyme and thus was also competitively inhibited by its product, galactose, at application-relevant concentrations. In order to alleviate this product inhibition, a model of the enzyme structure was generated based on a Penicillium sp. β-galactosidase crystal structure with bound β-galactose. This led to targeted mutagenesis of an Asp258-Ser-Tyr-Pro-Leu-Gly-Phe amino acid motif in the A. niger van Tiegh enzyme and isolation from the resultant library of a mutant β-galactosidase enzyme with reduced sensitivity to inhibition by galactose (K i of 6.46 mM galactose, compared with 0.76 mM for the wildtype recombinant enzyme). The mutated enzyme also exhibited an increased K m (3.76 mM compared to 2.21 mM) and reduced V max (110.8 μmol min−1 mg−1 compared to 172.6 μmol min−1 mg−1) relative to the wild-type enzyme, however, and its stability under simulated fasting gastric conditions was significantly reduced. The study nevertheless demonstrates the potential to rationally engineer the A. niger van Tiegh enzyme to relieve product inhibition and create mutants with improved, application-relevant kinetic properties for treatment of lactose intolerance.






Similar content being viewed by others
References
Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: signalP 3.0. J Mol Biol 340:783–795
Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, Phan I, Pilbout S, Schneider M (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res 31:365–370
Gaska J (1990) Treatment of lactose intolerance. Am Drug 202:36–43
Holsinger V (1988) Lactose. In: Wong P (ed) Fundamentals of dairy chemistry. Van Nostrand Reinhold, NY, pp 279–341
Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR (1989) Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61–68
Hu X, O’Dwyer R, Wall JG (2005) Cloning, expression and characterisation of a single-chain Fv antibody fragment against domoic acid in Escherichia coli. J Biotechnol 120:38–45
Hu X, O’Hara L, White S, Magner E, Kane M, Wall JG (2007) Optimisation of production of a domoic acid-binding scFv antibody fragment in Escherichia coli using molecular chaperones and functional immobilisation on a mesoporous silicate support. Protein Expr Purif 52:194–201
Jurado E, Camacho F, Luzon G, Vicaria JM (2002) A new kinetic model proposed for enzymatic hydrolysis of lactose by a β-galactosidase from Kluyveromyces fragilis. Enzyme Microb Technol 31:300–309
Kai Y, Kashiwagi T, Ishikawa K, Ziyatdinov MK, Redkina EI, Kiriukhin MY, Gusyatiner MM, Kobayashi S, Takagi H, Suzuki E (2006) Engineering of Escherichia coli L-serine O-acetyltransferase on the basis of crystal structure: desensitization to feedback inhibition by L-cysteine. Protein Eng Des Sel 19:163–167
Koganesawa N, Aizava T, Shimojo H, Miura K, Ohnishi A, Demura M, Hayakawa Y, Nitta K, Kawano K (2002) Expression and purification of a small cytokine growth-blocking peptide from armyworm Pseudaletia separata by an optimized fermentation method using the methylotrophic yeast Pichia pastoris. Protein Expr Purif 25:416–425
Labrou NE (2010) Random mutagenesis methods for in vitro directed enzyme evolution. Curr Protein Pept Sci 11:91–100
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) ClustalW2 and ClustalX version 2. Bioinformatics 23:2947–2948
Lin MY, Dipalma JA, Martini MC, Gross CJ, Harlander SK, Savaiano DA (1993) Comparative effects of exogenous lactase (β-galactosidase) preparations on in vivo lactose digestion. Dig Dis Sci 38:2022–2027
Macauley-Patrick S, Fazenda ML, McNeil B, Harvey LM (2005) Heterologous protein production using the Pichia pastoris expression system. Yeast 22:249–270
Nakayama T, Amachi T (1999) β-Galactosidase. In: Flinckinger MC, Drew SW (eds) Encyclopedia of bioprocess technology, fermentation, biocatalysis and bioseparation, 1st edn. Wiley, NY, pp 1291–1305
O’Connell S, Walsh G (2006) Physicochemical characteristics of commercial lactases relevant to their application in the alleviation of lactose intolerance. Appl Biochem Biotechnol 134:179–191
O’Connell S, Walsh G (2007) Purification and properties of a β-galactosidase with potential application as a digestive supplement. Appl Biochem Biotechnol 141:1–14
O’Connell S, Walsh G (2010) A novel acid-stable, acid-active β-galactosidase potentially suited to the alleviation of lactose intolerance. Appl Microbiol Biotechnol 86:517–524
Ogawa-Miyata Y, Kojima H, Sano K (2001) Mutation analysis of the feedback inhibition site of aspartokinase III of Escherichia coli K-12 and its use in L-threonine production. Biosci Biotechnol Biochem 65:1149–1154
Pollastri G, McLysaght A (2005) Porter: a new, accurate server for protein secondary structure prediction. Bioinformatics 21:1719–1720
Ramirez F, Lee K, Graham D (1994) All lactase preparations are not the same: results of a prospective, randomized, placebo-controlled trial. Am J Gastroenterol 89:566–570
Rasouli I, Kulkarni PR (1994) Enhancement of β-galactosidase productivity of Aspergillus niger NCIM-616. J Appl Bacteriol 77:359–361
Rojas AL, Nagem RA, Neustroev KN, Arand M, Adamska M, Eneyskaya EV, Kulminskaya AA, Garratt RC, Golubev AM, Polikarpov I (2004) Crystal structures of β-galactosidase from Penicillium sp. and its complex with galactose. J Mol Biol 343:1281–1292
Sambrook J, Russell DW (2001) Molecular cloning. A laboratory manual, 3rd edn. Cold Spring Harbor Laboratory, New York
Sanders S, Tolman K, Reitberg D (1992) Effect of a single dose of lactase on symptoms and expired hydrogen after lactose challenge in lactose-intolerant subjects. Clin Pharm 11:533–538
Takagi H, Kobayashi C, Kobayashi S, Nakamori S (1999) PCR random mutagenesis into Escherichia coli serine acetyltransferase: isolation of the mutant enzymes that cause overproduction of L-cysteine and L-cystine due to the desensitization to feedback inhibition. FEBS Lett 452:323–327
Acknowledgements
This study was funded by grant PC/2006/25 from Enterprise Ireland Science and Technology Development Agency.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hu, X., Robin, S., O’Connell, S. et al. Engineering of a fungal β-galactosidase to remove product inhibition by galactose. Appl Microbiol Biotechnol 87, 1773–1782 (2010). https://doi.org/10.1007/s00253-010-2662-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-010-2662-8