Abstract
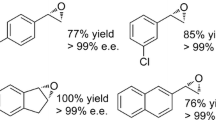
A gene encoding a putative epoxide hydrolase (EHase) was identified by analyzing an open reading frame of the genome sequence of Novosphingobium aromaticivorans, retaining the conserved catalytic residues such as the catalytic triad (Asp177, Glu328, and His355) and the oxyanion hole. The enantioselective EHase gene (neh) was cloned, and the recombinant EHase could be purified to apparent homogeneity by one step of metal affinity chromatography and further characterized. The purified N. aromaticivorans enantioselective epoxide hydrolase (NEH) showed enantioselective hydrolysis toward styrene oxide, glycidyl phenyl ether, epoxybutane, and epichlorohydrin. The optimal EHase activity toward styrene oxide occurred at pH 6.5 and 45°C. The purified NEH could preferentially hydrolyze (R)-styrene oxide with enantiomeric excess of more than 99% and 11.7% yield after 20-min incubation at an optimal condition. The enantioselective hydrolysis of styrene oxide was also confirmed by the analysis of the vicinal diol, 1-phenyl-1,2-ethanediol. The hydrolyzing rates of the purified NEH toward epoxide substrates were not affected by as high as 100 mM racemic styrene oxide.







Similar content being viewed by others
References
Arahira M, Nong VH, Udaka K, Fukazawa C (2000) Purification, molecular cloning and ethylene-inducible expression of a soluble-type epoxide hydrolase from soybean (Glycine max [L.] Merr.). Eur J Biochem 267:2649–2657
Arand M, Wagner H, Oesch F (1996) Asp333, Asp495, and His523 form the catalytic triad of rat soluble epoxide hydrolase. J Biol Chem 271:4223–4229
Arand M, Hemmer H, Durk H, Baratti J, Archelas A, Furstoss R, Oesch F (1999) Cloning and molecular characterization of a soluble epoxide hydrolase from Aspergillus niger that is related to mammalian microsomal epoxide hydrolase. Biochem J 344:273–280
Archelas A, Furstoss R (1997) Synthesis of enantiopure epoxides through biocatalytic approaches. Annu Rev Microbiol 51:491–525
Archelas A, Furstoss R (2001) Synthetic applications of epoxide hydrolases. Curr Opin Chem Biol 5:112–119
Bhatnagar T, Manoj KM, Baratti JC (2001) A spectrophotometric method to assay epoxide hydrolase activity. J Biochem Biophys Methods 50:1–13
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Cao L, Lee J, Chen W, Wood TK (2006) Enantioconvergent production of (R)-1-phenyl-1,2-ethanediol from styrene oxide by combining the Solanum tuberosum and an evolved Agrobacterium radiobacter AD1 epoxide hydrolases. Biotechnol Bioeng 94:522–529
Chen C-S, Fujimoto Y, Girdaukas G, Sih CJ (1982) Quantitative analyses of biochemical kinetic resolutions of enantiomers. J Am Chem Soc 104:7294–7299
Falany CN, McQuiddy P, Kasper CB (1987) Structure and organization of the microsomal xenobiotic epoxide hydrolase gene. J Biol Chem 262:5924–5930
Hwang Y-O, Kang SG, Woo J-H, Kwon KK, Sato T, Lee EY, Han MS, Kim S-J (2008) Screening enantioselective epoxide hydrolase activities from marine microorganisms: detection of activities in Erythrobacter spp. Marine Biotechnol 10:366–373
Janssen DB, Pries F, van der Ploeg J, Kazemier B, Terpstra P, Witholt B (1989) Cloning of 1,2-dichloroethane degradation genes of Xanthobacter autotrophicus GJ10 and expression and sequencing of the dhlA gene. J Bacteriol 171:6791–6799
Kim JT, Kang SG, Woo J-H, Lee J-H, Jeong BC, Kim S-J (2007) Screening and its potential application of lipolytic activity from a marine environment: characterization of a novel esterase from Yarrowia lipolytica CL180. Appl Microbiol Bioterchnol 74:820–828
Knehr M, Thomas H, Arand M, Gebel T, Zeller HD, Oesch F (1993) Isolation and characterization of a cDNA encoding rat liver cytosolic epoxide hydrolase and its functional expression in Escherichia coli. J Biol Chem 268:17623–17627
Kumar S, Tamura K, Nei M (2004) MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the heal of bacteriophage T4. Nature 227:680–685
Lee EY, Shuler ML (2007) Molecular engineering of epoxide hydrolase and its application to asymmetric and enantioconvergent hydrolysis. Biotechnol Bioeng 98:318–327
Misawa E, Chion CK, Archer IV, Woodland MP, Zhou NY, Carter SF, Widdowson DA, Leak DJ (1998) Characterisation of a catabolic epoxide hydrolase from a Corynebacterium sp. Eur J Biochem 253:173–183
Nardini M, Dijkstra BW (1999) α/β hydrolase fold enzymes: the family keeps growing. Curr Opin Struct Biol 9:732–737
Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J, Sussman JL, Verschueren KHG, Goldman A (1992) The α/β hydrolase fold. Protein Eng 5:197–211
Oppenheimer CH, ZoBell CE (1952) The growth and viability of sixty-three species of marine bacteria as influenced by hydrostatic pressure. J Mar Res 11:10–18
Park S-Y, Kim J-T, Kang SG, Woo J-H, Lee J-H, Choi H-T, Kim S-J (2007) A new esterase showing similarity to putative dienelactone hydrolase from a strict marine bacterium, Vibrio sp. GMD509. Appl Microbiol Bioterchnol 77:107–115
Rink R, Fennema M, Smids M, Dehmel U, Janssen DB (1997) Primary structure and catalytic mechanism of the epoxide hydrolase from Agrobacterium radiobacter AD1. J Biol Chem 272:14650–14657
Rink R, Spelberg JHL, Pieters RJ, Kingma J, Nardini M, Kellogg RM, Dijkstra BW, Janssen DB (1999) Mutation of tyrosine residues involved in the alkylation half reaction of epoxide hydrolase from Agrobacterium radiobacter AD1 results in improved enantioselectivity. J Am Chem Soc 121:7417–7418
Rink R, Kingma J, Spelberg JH, Janssen DB (2000) Tyrosine residues serve as proton donor in the catalytic mechanism of epoxide hydrolase from Agrobacterium radiobacter. Biochemistry 39:5600–5613
Rui L, Cao L, Chen W, Reardon KF, Wood TK (2005) Protein engineering of epoxide hydrolase from Agrobacterium radiobacter AD1 for enhanced activity and enantioselective production of (R)-1-phenylethane-1,2-diol. Appl Environ Microbiol 71:3995–4003
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn, vol 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 8.65–8.71
Stapleton A, Beetham JK, Pinot F, Garbarino JE, Rockhold DR, Friedman M, Hammock BD, Belknap WR (1994) Cloning and expression of soluble epoxide hydrolase from potato. Plant J 6:251–258
Straathof AJJ, Jongejan JA (1997) The enantiomeric ratio: origin, determination and prediction. Enzyme Microb Tech 21:559–571
Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, Zeeberg B, Buetow KH, Schaefer CF, Bhat NK, Hopkins RF, Jordan H, Moore T, Max SI, Wang J, Hsieh F, Diatchenko L, Marusina K, Farmer A, Rubin GM, Hong L, Stapleton M, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, Brownstein MJ, Usdin TB, Toshiyuki S, Carninci P, Prange C, Raha SS, Loquellano NA, Peters GJ, Abramson RD, Mullahy SJ, Bosak SA, McEwan PJ, McKernan KJ, Malek JA, Gunaratne PH, Richards S, Worley KC, Hale S, Garcia AM, Gay LJ, Hulyk SW, Villalon DK, Muzny DM, Sodergren EJ, Lu X, Gibbs RA, Fahey J, Helton E, Ketteman M, Madan A, Rodrigues S, Sanchez A, Whiting M, Madan A, Young AC, Shevchenko Y, Bouffard GG, Blakesley RW, Touchman JW, Green ED, Dickson MC, Rodriguez AC, Grimwood J, Schmutz J, Myers RM, Butterfield YS, Krzywinski MI, Skalska U, Smailus DE, Schnerch A, Schein JE, Jones SJ, Marra MA (2002) Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA 99:16899–16903
Thompson JD, Higgin DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tokunaga M, Larrow JF, Kakiuchi F, Jacobsen EN (1997) Asymmetric catalysis with water: efficient kinetic resolution of terminal epoxides by means of catalytic hydrolysis. Science 277:936–938
van Loo B, Spelberg JHL, Kingma J, Sonke T, Wubbolts MG, Janssen DB (2004) Directed evolution of epoxide hydrolase from A. radiobacter toward higher enantioselectivity by error-prone PCR and DNA shuffling. Chem Biol 11:981–990
van Loo B, Kingma J, Arand M, Wubbolts MG, Janssen DB (2006) Diversity and biocatalytic potential of epoxide hydrolases identified by genome analysis. Appl Envir Microbiol 72:2905–2917
Visser H, de Bont JAM, Verdoes JC (1999) Isolation and characterization of the epoxide hydrolase-encoding gene from Xanthophyllomyces dendrorhous. Appl Environ Microbiol 65:5459–5463
Visser H, Vreugdenhil S, de Bont JAM, Verdoes JC (2000) Cloning and characterization of an epoxide hydrolase-encoding gene from Rhodotorula glutinis. Appl Microbiol Biotechnol 53:415–419
Weijers CAGM, de Bont JAM (1999) Epoxide hydrolases from yeasts and other sources: versatile tools in biocatalysis. J Mol Catal B Enzym 6:199–214
Woo J-H, Hwang O-K, Kang SG, Lee HS, Cho J, Kim S-J (2007) Cloning and characterization of three novel epoxide hydrolases from a marine bacterium, Erythrobacter litoralis HTCC2594. Appl Microbiol Biotechnol 76:365–375
Acknowledgment
This work was supported by KORDI in-house program (PE98202) and the Marine and Extreme Genome Research Center Program, Ministry of Marine Affairs and Fisheries, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Woo, JH., Kang, JH., Kang, S.G. et al. Cloning and characterization of an epoxide hydrolase from Novosphingobium aromaticivorans . Appl Microbiol Biotechnol 82, 873–881 (2009). https://doi.org/10.1007/s00253-008-1791-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-008-1791-9