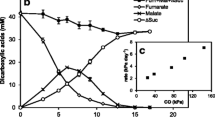
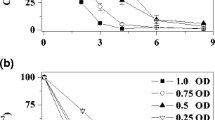
Abstract
Shewanella decolorationis S12, a representative dissimilatory azo-reducing bacterium of Shewanella genus, can grow by coupling the oxidation of hydrogen to the reduction of azo compounds as the sole electron acceptor, indicating that an uptake hydrogenase is an important component for electron transfer for azoreduction. For searching to the uptake hydrogenase in the genome of S. decolorationis, two operons, hyd and hya, were cloned and sequenced, which encode periplasmically oriented Fe-only hydrogenase and a Ni–Fe hydrogenase, respectively, according to the homologous comparison with other bacterial hydrogenases. In order to assess the roles of these two enzymes in hydrogen-dependent azoreduction and growth, hyd- and hya-deficient mutants were generated by gene replacement. Hya was found to be required for hydrogen-dependent reduction of azo compound by resting cell suspensions and to be essential for growth with hydrogen as electron donor and azo compound as electron acceptor. Hyd, in contrast, was not. These findings suggest that Hya is an essential respiratory hydrogenase of dissimilatory azoreduction in S. decolorationis.




Similar content being viewed by others
References
Adams MWW, Mortenson LE, Chen JS (1981) Hydrogenase. Biochim Biophys Acta 594:105–176
Blumel S, Knackmuss H-J, Stolz A (2002) Molecular cloning and characterization of the gene coding for the aerobic azoreductase from Xenophilus azovorans KF46F. Appl Environ Microbiol 68:3948–3955
Blumel S, Stolz A (2003) cloning and characterization of the gene coding for the aerobic azoreductase from Pigmentiphaga kullae K24. Appl Microbiol Biotechnol 62:186–190
Brigé A, Motte B, Borloo J, Buysschaert G, Devreese B, Van Beeumen JJ (2008) Bacterial decolorization of textile dyes is an extracellular process requiring a multicomponent electron transfer pathway. Microb Biotechnol 1:40–52
Brown MA, DeVito SC (1993) Predicting azo dye toxicity. Crit Rev Environ Sci Technol 23:249–324
Chen H, Wang R-F, Cerniglia CE (2004) Molecular cloning, over expression, purification, and characterization of an aerobic FMN-dependent azoreductase from Enterococcus faecalis. Protein Expr Purif 34:302–310
Chung KT, Cerniglia CE (1992) Mutagenicity of azo dyes: structure–activity relationships. Mutat Res 77:201–220
Dennis JJ, Zylstra GJ (1998) Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl Environ Microbiol 64:2710–2715
Dross F, Geisler V, Lenger R, Theis F, Krafft T, Fahrenholz F, Kojro E, Duchêne A, Tripier D, Juvenal K, Kröger A (1992) The quinone-reactive Ni/Fe-hydrogenase of Wolinella succinogenes. Eur J Biochem 206:93–102
Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 136:557–580
Hanahan D (1985) Techniques for transformation of Escherichia coli. In: Glover DM (ed) DNA cloning, vol I, a practical approach. IRL, Oxford, pp 109–135
Heidelberg JF, Paulsen IT, Nelson KE, Gaidos EJ, Nelson WC, Read TD, Eisen JA, Seshadri R, Ward N, Methe B, Clayton RA, Meyer T, Tsapin A, Scott J, Beanan M, Brinkac L, Daugherty S, DeBoy RT, Dodson RJ, Durkin AS, Haft DH, Kolonay JF, Madupu R, Peterson JD, Umayam LA, White O, Wolf AM, Vamathevan J, Weidman J, Impraim M, Lee K, Berry K, Lee C, Mueller J, Khouri H, Gill J, Utterback TR, McDonald LA, Feldblyum TV, Smith HO, Venter JC, Nealson KH, Fraser CM (2002) Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat Biotechnol 20:1118–1123
Hidalgo E, Leyva A, Ruiz-Argüeso T (1990) Nucleotide sequence of the hydrogenase structural genes from Rhizobium leguminosarum. Plant Mol Biol 15:367–370
Higuchi Y, Yagi T, Yasouka N (1997) Unusual ligand structure in Ni–Fe active center and an additional Mg site in hydrogenase revealed by high resolution X-ray structure analysis. Structure 5:1671–1680
Hong Y, Xu M, Guo J, Xu Z, Chen X, Sun G (2007) Respiration and growth of Shewanella decolorationis S12 with azo compound as sole electron acceptor. Appl Environ Microbiol 73:64–72
Kudlich M, Keck A, Klein J, Stolz A (1997) Localization of the enzyme system involved in anaerobic reduction of azo dyes by Sphingomonas sp. strain BN6 and effect of artificial redox mediators on the rate of azo dye reduction. Appl Environ Microbiol 63:3691–3694
Lovley DR, Coates JD, Blunt-Harris EL, Phillips EJP, Woodward JC (1996) Humic substances as electron acceptors for microbial respiration. Nature 382:441–448
Lovley DR, Phillips EJP, Gorby YA, Landa ER (1991) Microbial reduction of uranium. Nature 350:413–416
Marshall MJ, Plymale AE, Kennedy DW, Shi L, Wang Z, Reed SB, Dohnalkova AC, Simonson CJ, Liu C, Saffarini DA, Romine MF, Zachara JM, Beliaev AS, Fredrickson JK (2008) Hydrogenase- and outer membrane c-type cytochrome-facilitated reduction of technetium(VII) by Shewanella oneidensis MR-1. Environ Microbiol 10:125–136
Menon NK, Robbins J, Peck HD, Chatelus CY, Choi ES, Przybyla AE (1990) Cloning and sequencing of a putative Escherichia coli [Ni–Fe] hydrogenase-1 operon containing six open reading frames. J Bacteriol 172:1969–1977
Meshulam-Simon G, Behrens S, Choo AD, Spormann AM (2007) Hydrogen metabolism in Shewanella oneidensis MR-1. Appl Environ Microbiol 73:1153–1165
Miller TL, Wolin ML (1974) A serum bottle modification of the Hungate technique for cultivating obligate anaerobes. Appl Microbiol 27:985–987
Myers CR, Myers JM (1994) Ferric iron reduction-linked growth yields of Shewanella putrefaciens MR-1. J Appl Bacteriol 76:253–258
Myers CR, Nealson KH (1998) Bacterial manganese reduction and growth with manganese oxide as sole electron acceptor. Science 240:1319–1321
Myers CR, Nealson KH (1990) Respiration-linked proton translocation coupled to anaerobic reduction of manganese(IV) and iron(III) in Shewanella putrefaciens MR-1. J Bacteriol 172:6232–6238
Nakanishi M, Yatome C, Ishida N, Kitade Y (2001) Putative ACP phosphodiesterase gene (acpD) encodes an azoreductase. J Biol Chem 276:46394–46399
Posewitz MC, King PW, Smolinski S Zhang LL, Seibert M, Ghirardi ML (2004) Discovery of two novel radical S-adenosylmethionine proteins required for the assembly of an active [Fe] hydrogenase. J Biol Chem 279:25711–25720
Prickril BC, Czechowski MH, Przybyla AE, Peck HD, LeGall J (1986) Putative signal peptide on the small subunit of the periplasmic hydrogenase from Desilfovibrio vulgaris. J Bacteriol 167:722–725
Rafii F, Franklin W, Cerniglia CE (1990) Azoreductase activity of anaerobic bacteria isolated from human intestinal microflora. Appl Environ Microbiol 56:2146–2151
Rau J, Stolz A (2003) Oxygen-insensitive nitroreductases NfsA and NfsB of Escherichia coli function under anaerobic conditions as lawsone-dependent azo reductases. Appl Environ Microbiol 69:3448–3455
Sambrook J, Fritsch EF, Manistis T (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York
Simon R, Priefer U, Pühler A (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:784–791
Stolz A (2001) Basic and applied aspects in the microbial degradation of azo dyes. Appl Microbiol Biotechnol 56:69–80
Suzuki Y, Yoda T, Ruhul A, Sugiura W (2001) Molecular cloning and characterization of the gene coding for azoreductase from Bacillus sp. OY1-2 isolated from soil. J Biol Chem 276:9059–9065
Voordouw G, Brenner S (1985) Nucleotide sequence of the gene encoding the hydrogenase from Desulfovibrio vulgaris (Hildenborough). Eur J Biochem 148:515–520
Voordouw G, Strang JD, Wilson FR (1989) Organization of the genes encoding [Fe] hydrogenase in Desulfovibrio vulgaris subsp. oxamicus Monticello. J Bacteriol 171:3881–3889
Volbeda A, Charon MH, Piras C, Hatchikian EC, Frey M, FontecillaCamps JC (1995) Crystal structure of the nickel–iron hydrogenase from Desulfovibrio gigas. Nature 373:580–587
Volbeda A, Garcin E, Piras C, de Lacey AL, Fernandez VM, Hatchikian EC, Frey M, FontecillaCamps JC (1996) Structure of the [Ni–Fe] hydrogenase active site: evidence for biologically uncommon Fe ligands. J Am Chem Soc 118:12989–12996
Von Heijne G (1985) Signal sequences: the limits of variation. J Mol Biol 184:99–105
Xu M, Guo J, Cen Y, Zhong X, Cao W, Sun G (2005) Shewanella decolorationis sp. nov., a dye-decolorizing bacterium isolated from an activated sludge of waste-water treatment plant. Int J Syst Evol Microbiol 55:363–368
Acknowledgments
The author gratefully acknowledges the support of Young People Innovation Foundation of South China Sea Institute of Oceanology, CAS (07SC011009) and National Natural Science Foundation (30670020).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1.
Nucleic acid sequence of Fe hydrogenase from S. decolorationnis S12. The coding regions of the genes for the 46-kDa hydrogenase (169-1401) and the 13.5-kDa protein (1414-1734) are indicated by means of translocation using the single-letter amino acid code. The Cys residues have been circled. Two possible ribosome binding sites, as well as a promoter, are indicated (−35, −10). (DOC 29.5 KB)
Fig. S2.
Nucleic acid sequence of Ni–Fe hydrogenase from S. decolorationnis S12. The coding regions of the genes for the small subunit (136-1272), large subunit (1276-2976), cytochrome b subunit (2998-3675) are indicated by means of translocation using the single-letter amino acid code. The Cys and His residues have been circled. Three possible ribosome binding sites, as well as a promoter, are indicated (−35, −10). (DOC 42.0 KB)
Rights and permissions
About this article
Cite this article
Hong, YG., Guo, J. & Sun, GP. Identification of an uptake hydrogenase for hydrogen-dependent dissimilatory azoreduction by Shewanella decolorationis S12. Appl Microbiol Biotechnol 80, 517–524 (2008). https://doi.org/10.1007/s00253-008-1597-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-008-1597-9