Abstract
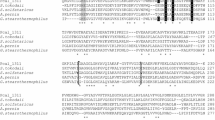
Alcohol dehydrogenase (ADH) is a key enzyme in the production and utilization of alcohols. Some also catalyze the formation of carboxylate esters from alcohols and aldehydes. The ADH1 and ADH3 genes of Neurospora crassa FGSC2489 were cloned and expressed in recombinant Escherichia coli to investigate their alcohol dehydrogenation and carboxylate ester formation abilities. Homology analysis and sequence alignment of amino acid sequence indicated that ADH1 and ADH3 of N. crassa contained a zinc-binding consensus sequence and a NAD+-binding motif and showed 54–75% identity with fungi ADHs. N. crassa ADH1 was expressed in E. coli to give a specific activity of 289 ± 9 mU/mg using ethanol and NAD+ as substrate and cofactor, respectively. Corresponding experiments on the expression and activity of ADH3 gave 4 mU/mg of specific activity. N. crassaADH1 preferred primary alcohols containing C3–C8 carbons to secondary alcohols such as 2-propanol and 2-butanol. N. crassaADH1 possessed 5.3 mU/mg of specific carboxylate ester-forming activity accumulating 0.4 mM of ethyl acetate in 18 h. Substrate specificity of various linear alcohols and aldehydes indicated that short chain-length alcohols and aldehydes were good substrates for carboxylate ester production. N. crassaADH1 was a primary alcohol dehydrogenase using cofactor NAD+ preferably and possessed carboxylate ester-forming activity with short chain alcohols and aldehydes.





Similar content being viewed by others
References
Bruchez JJP, Eberle J, Kohler W, Kruft V, Radford A, Russo VEA (1996) bli-4, a gene that is rapidly induced by blue light, encodes a novel mitochondrial, short-chain alcohol dehydrogenase-like protein in Neurospora crassa. Mol Gen Genet 252:223–229
Galagan JE, Calvo SF, Borkovich KA, Selke, EU, Read ND, Jaffe D, FitzHugh W, Ma LJ, Smirnov S, Purcell S, Rehman B, Elkins T et al (2003) The genome sequence of the filamentous fungus Neurospora crassa. Nature 422:859–868
Horton CE, Huang KX, Bennett GN, Rudolph FB (2003) Heterologous expression of the Saccharomyces cerevisiae alcohol acetyltransferase genes in Clostridium acetobutylicum and Escherichia coli for the production of isoamyl acetate. J Ind Microbiol Biotech 30:427–432
Kim SG, Kweon DH, Lee DH, Park YC, Seo JH (2005) Coexpression of folding accessory proteins for production of active cyclodextrin glycosyltransferase of Bacillus macerans in recombinant Escherichia coli. Protein Expr Purif 41:426–432
Kotani T, Yamamoto T, Yurimoto H, Sakai Y, Kato N (2003) Propane monooxygenase and NAD+-dependent secondary alcohol dehydrogenase in propane metabolism by Gordonia sp. Strain TY-5. J Bacteriol 185:7120–7128
Kusano M, Sakai Y, Kato N, Yoshimoto H, Sone H, Tamai Y (1998) Hemiacetal dehydrogenation activity of alcohol dehydrogenases in Saccharomyces cerevisiae. Biosci Biotechnol Biochem 62:1956–1961
Kusano M, Sakai Y, Kato N, Yoshimoto H, Tamai Y (1999) A novel hemiacetal dehydrogenase activity involved in ethyl acetate synthesis in Candida utilis. J Biosci Bioeng 87:690–692
Leskovac V, Trivić S, Peričin D (2002) The three zinc-containing alcohol dehydrogenases from baker’s yeast, Saccharomyces cerevisiae. FEMS Yeast Res 2:481–494
Murdanoto AP, Sakai Y, Sembiring L, Tani Y, Kato N (1997) Ester synthesis by NAD+-dependent dehydrogenation of hemiacetal: production of methyl formate by cells of methylotrophic yeasts. Biosci Biotech Biochem 61:1391–1393
Park YC, Yun NR, San KY, Bennett GN (2006) Molecular cloning and characterization of the alcohol dehydrogenase ADH1 gene of Candida utilis ATCC 9950. J Ind Microbiol Biotech 33:1032–1036
Pastore GM, Park YK, Min DB (1994) Production of fruity aroma by Neurospora from beiju. Mycol Res 98:1300–1302
Reid MR, Fewson CA (1994) Molecular characterization of microbial alcohol dehydrogenase. Crit Rev Microbiol 20:13–56
Sakai Y, Murdanoto AP, Sembiring L, Tani Y, Kato N (1995) A novel formaldehyde oxidation pathway in methylotrophic yeasts: methylformate as a possible intermediate. FEMS Microbiol Lett 127:229–234
Verduyn C, Breedveld GJ, Scheffers WA, Van Dijken JP (1988) Substrate specificity of alcohol dehydrogenase from the yeasts Hansenula polymorpha CBS 4732 and Candida utilis CBS 621. Yeast 4:143–148
Xie X, Wilkinson HH, Correa A, Lewis ZA, Bell-Pedersen D, Ebbole DJ (2004) Transcriptional response to glucose starvation and functional analysis of a glucose transporter of Neurospora crassa. Fungal Genet Biol 41:1104–1119
Yoshizawa K, Yamauchi H, Hasuo T, Akita O, Hara S (1988) Production of fruity odor by Neurospora sp. Agric Biol Chem 52:2129–2130
Young ET, Pilgrim D (1985) Isolation and DNA sequence of ADH3, a nuclear gene encoding the mitochondrial isozyme of alcohol dehydrogenase in Saccharomyces cerevisiae. Mol Cell Biol 5:3024–3034
Yurimoto H, Lee B, Yasuda F, Sakai Y, Kato N (2004) Alcohol dehydrogenases that catalyse methyl formate synthesis participate in formaldehyde detoxification in the methylotrophic yeast Candida boidinii. Yeast 21:341–350
Acknowledgment
This study was supported by the Korea Research Foundation Grant (KRF-2004-000-10255-0) and USDA-NSF Interagency Metabolic Engineering Program: USDA 2002-35505-11638 and NSF BES 0118815.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Park, YC., San, KY. & Bennett, G.N. Characterization of alcohol dehydrogenase 1 and 3 from Neurospora crassa FGSC2489. Appl Microbiol Biotechnol 76, 349–356 (2007). https://doi.org/10.1007/s00253-007-0998-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-007-0998-5