Abstract
In comparison to other bacteria Bacillus subtilis emits the volatile compound isoprene in high concentrations. Isoprene is the smallest representative of the natural product group of terpenoids. A search in the genome of B. subtilis resulted in a set of genes with yet unknown function, but putatively involved in the methylerythritol phosphate (MEP) pathway to isoprene. Further identification of these genes would give the possibility to engineer B. subtilis as a host cell for the production of terpenoids like the valuable plant-produced drugs artemisinin and paclitaxel. Conditional knock-out strains of putative genes were analyzed for the amount of isoprene emitted. Differences in isoprene emission were used to identify the function of the enzymes and of the corresponding selected genes in the MEP pathway. We give proof on a biochemical level that several of these selected genes from this species are involved in isoprene biosynthesis. This opens the possibilities to investigate the physiological function of isoprene emission and to increase the endogenous flux to the terpenoid precursors, isopentenyl diphosphate and dimethylallyl diphosphate, for the heterologous production of more complex terpenoids in B. subtilis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Isoprene (2-methyl-1,3-butadiene; I) or actually isopentenyl diphosphate (IDP; II) is the general precursor of all terpenoids, that represent a very diverse class of natural products. Two evolutionary distinct routes occur in nature for the biosynthesis of IDP. The spread of the two pathways is well investigated for organisms with a sequenced genome (Lange et al. 2000; Boucher and Doolittle 2000; Rohdich et al. 2001). In eukaryotes and archaea IDP and its isomer dimethylallyl diphosphate (DMADP; III) are formed via the mevalonate pathway (reviewed by Kuzuyama 2002). This pathway is well studied and for many organisms the enzymes are characterized and the encoding genes identified. More recently, another pathway to IDP was discovered in some eubacteria and in plastids of higher plants, that proceeds via the intermediate methylerythritol phosphate (MEP) (Rohmer et al. 1993; Rohmer 1999).
Most Gram-negative bacteria, including Escherichia coli, use the MEP pathway and for E. coli this is now well documented. For Gram-positive bacteria the situation is less clear. Gram-positive cocci have been reported to only possess genes for the mevalonate pathway (Wilding et al. 2000). In Streptomyces aeriouvifer both pathways are found (Seto et al. 1996). In Bacillus subtilis, being regarded as the prototype for Gram-positives, only homologues of the genes for the MEP pathway are present (Wagner et al. 2000).
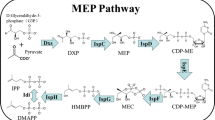
For E. coli the complete MEP pathway has been elucidated and the genes involved have been identified and the corresponding enzymes described (reviewed by Eisenreich et al. 2004). The pathway exists of seven subsequent enzymatic steps (Fig. 1). The first reaction of the MEP pathway is catalyzed by a transketolase (DXS) and concerns the condensation of pyruvate (IV) and d-glyceraldehyde-3-phosphate (V) to 1-deoxy-d-xylulose 5-phosphate (VI) (Sprenger et al. 1997; Lois et al. 1998). 1-deoxy-d-xylulose is a branching intermediate in the biosynthesis of isoprenoids, thiamine (vitamin B1) (White 1978; David et al. 1981, 1982), and pyridoxol (vitamin B6), respectively (Hill et al. 1989). The second step is catalyzed by the IspC protein, initiating a rearrangement in the carbon skeleton followed by a reduction in the keto function of (VI), and finally delivering 2C-methyl-d-erythritol 4-phosphate (Takahashi et al. 1998). Five other subsequently acting enzymes are needed for the synthesis of IDP, including a phosphocytidyl transferase (IspD), a kinase (IspE), a cyclophosphate synthase (IspF), and two reductases (IspG, IspH) (Fig. 1).
Methylerythritol phosphate pathway of isoprenoids [Dxs (1), 1 deoxy-d-xylulose 5-phosphate synthase; IspC (2), 2C-methyl-d-erythritol 4-phosphate synthase; IspD (3), 4-diphosphocytidyl-2C-methyl-d-erythritol 4-phosphate synthase; IspE (4), 4-diphosphocytidyl-2C-methyl-d-erythritol kinase; IspF (5), 2C-methyl-d-erythritol 2,4-diphosphate synthase; IspG (6), 2C-methyl-d-erythritol 2,4-cyclodiphosphate; IspH (7), 1-hydroxy-2-methyl-butenyl 4-diphosphate reductase; Idi (8), isopentyl diphosphate isomerase; IspA (9), farnesyl diphosphate synthase]. The putative genes are mentioned in italics for every biosynthetic step, including the essential gene YpgA, which is indicated with an *. Numbering of compounds and biosynthetic steps refers to the text
Bacteria use the precursors IDP and DMADP for the synthesis of several compounds including the side chains of ubiquinone or menaquinone. Next to that, several bacterial species use these precursors to synthesize isoprene, that is emitted to their environment (Kuzma et al. 1995). B. subtilis emits isoprene in high levels compared to other bacterial species as has been described for B. subtilis 6051, B. subtilis 23059, and B. subtilis 23856 (Kuzma et al. 1995). Using a bioreactor system, the emission of isoprene by B. subtilis 6051 was found to occur in three phases during the growth curve (Wagner et al. 1999). The phases correspond with respectively, glucose metabolism and secretion of acetoin, catabolism of acetoin, and the early stage of sporulation. The general laboratory strain B. subtilis 168 showed another pattern of isoprene emission, lacking phases 2 and 3 (Fall and Copley 2000). The function of the emitted isoprene has been postulated as being a signal molecule in the natural environment of the microorganism. Another possible explanation for the emitted isoprene is the efflux as an overflow metabolite in the bacterial pathway to isoprenoid structures (Fall and Copley 2000). The uptake of isoprene by microorganisms present in soil samples has been described as a sink of atmospheric isoprene (Cleveland and Yavitt 1998). However, there is no full evidence supporting one of the hypotheses mentioned. Isoprene originates from DMADP. The conversion of DMADP in isoprene is known to be an enzymatic process in the poplar tree from which a gene has been isolated and characterized (Miller et al. 2001; Schnitzler et al. 2005). Attempts to prove the enzymatic conversion to isoprene in B. subtilis suggested the involvement of an enzymatic step. Enzymatic activity has been partially purified, but the enzyme turned out to be very labile (Sivy et al. 2002). Until now the encoding gene could not be identified. A search for a homologue protein of the isoprene synthase from poplar tree in the genome of the B. subtilis did not yield a candidate. Based on homology with known genes, mostly from E. coli, candidate genes for the other enzymatic steps in the MEP pathway in B. subtilis have been suggested. Identification of the genes involved in this pathway should shed some light on the function of isoprene synthesis in Bacillus physiology. Due to the essentiality of the products downstream of the isoprenoid biosynthesis, like menaquinone, it is not remarkable that the candidate genes, except for the isomerase, have been shown to be essential genes for the survival of the bacterial cell (Kobayashi et al. 2003). In the construction of the BFAN collection these knock-out strains were not viable and therefore conditional mutants have been made using a pMUTIN3-vector. The expression in these strains can be regulated by the isopropyl-beta-d-thiogalactopyranoside (IPTG) inducible Pspac promoter (Yansura and Henner 1984; Vagner et al. 1998). Using this mutant system, expression levels can be regulated to a minimum for survival by decreasing the amounts of IPTG. In that way, the function of the genes can be investigated. Identification of the involvement of certain genes in the isoprenoid biosynthetic pathway, may create possibilities to redirect the metabolic flux toward IDP. The possibilities of pathway engineering have shown their strength already in other microorganisms, like E. coli and Saccharomyces cerevisiae. The metabolic flux was dependent mainly on the deoxyxylulose 5-phosphate synthase (DXS), the isopentenyl diphosphate isomerase (Idi), and the prenyltransferase delivering farnesyl diphosphate (IspA) (Harker and Bramley 1999; Kim and Keasling 2001; Kajiwara et al. 1997; Martin et al. 2003). By up-regulation of the MEP pathway, B. subtilis can be developed as an interesting host organism for the production of complex terpenoid compounds, such as the valuable drugs artemisinin and paclitaxel. In the present study we describe the functional analysis of genes in B. subtilis putatively encoding enzymes involved in the biosynthesis of the terpenoid precursor isopentenyl diphosphate. We used quantitative isoprene emission of different well controllable knock-out Bacillus strains in their environment to determine the function of the tested genes.
Materials and methods
Bacterial strains and media
Bacterial strains used are listed in Table 1. B. subtilis strains 6051, 23059 and 23856 were obtained from the American Type Culture Collection (ATCC) (Rockville, USA). B. subtilis mutant strains were obtained through a chromosomal integration of pMUTIN3 derived plasmids (Vagner et al. 1998). Experiments were performed using Luria-Bertani (LB)-medium containing 1% bacto-tryptone, 0.5% yeast extract, and 0.5% NaCl (all purchased from Duchefa, Zwijndrecht, the Netherlands). If required, medium was supplemented with erythromycin (2 μg/ml; Duchefa, the Netherlands) or the specified concentration of isopropyl-β-d-thiogalactopyranoside (IPTG; Duchefa, the Netherlands). For growing on plates the medium was solidified with 1.5% agar.
Detection of bacterial isoprene emission
A single colony of the different B. subtilis strains was transferred from a plate to 10 ml Luria-Bertani (LB) medium (if required supplemented with 2 μg/ml erythromycin and 100 μM IPTG) and grown over night (37°C; 300 rpm). Before the inoculation (1:100) of fresh LB medium containing different specified concentrations of IPTG, cells were gently washed three times with fresh LB-medium without IPTG by resuspending and centrifugating.
Isoprene accumulation was measured on-line by sampling every 15 min, during a period of nine hours, 15 ml of the air above 50 ml bacterial culture, growing (37°C; 300 rpm) in a 500 ml Erlenmeyer air tight flask (CBN, the Netherlands), and transferring into a gas-chromatography system suitable for the sensitive detection of isoprene (Syntech Spectras GC955 series 601, Synspec BV, the Netherlands) (Loreto and Delfine 2000). The air was pumped through a Tenax GA trap, desorbed at 180°C and transferred to an AT 5 column under a flow of 2.5 ml/min nitrogen (3.7 bars; quality 5). The temperature program used was 3 min at 50°C followed by an increase in temperature to 70°C at 5 min; kept at this temperature until 12 min and than lowered to 50°C again. The isoprene present was detected by photo ionization at 10.6 eV. The gas chromatograph was calibrated using the dynamic gas dilution principle with several concentrations of gaseous isoprene using liquid isoprene (Sigma, USA) diluted in methanol and evaporated with a gas dilutor (MK5, MCZ Umwelttechnik, Germany). During the isoprene detection the growth of the bacterial culture was determined by measuring the optical density at 600 nm (OD600 nm) every hour.
Results
Isoprene emission in wild-type Bacillus strains
The emission of isoprene from B. subtilis wild type strains 168, 6051, 23059, and 23856 was investigated. For all four strains isoprene accumulated in the logarithmic phase of growth, leading to a high increase in isoprene in the flask. The absolute maximum of wild type strains varied between 400 and 700 μg/m3. Starting from the late logarithmic phase to the beginning of the stationary phase the amount of isoprene in the flask decreased slowly.
Figure 2 shows the accumulation of isoprene during the growth of the wild type strains 168 and 6051. B. subtilis 6051 reached higher levels of isoprene, but this is probably not caused by a higher production rate rather than the amount of cells present, as reflected by the optical density of these strains. Corrected for the amount of cells, determined as the optical density at 600 nm, B. subtilis 168 accumulated at maximum 148 μg/ m3 / OD600 nm compared to 166 μg/ m3 / OD600 nm for B. subtilis 6051. The other two wild type strains tested, 23059 and 23856, showed a similar accumulation of isoprene as the strains described before. Corrected for the amount of cells, 23059 produced 192 μg/ m3 / OD600 nm and 23856 produced 112 μg/ m3 / OD600 nm isoprene at maximum accumulation (Table 2).
Mutant strains
To find the optimal conditions in which expression of the genes can be limited without killing the cells, each conditional mutant strain was subjected to IPTG depletion by growing the strains on agar plates containing different concentrations of IPTG varying from 0 to 1 mM. Decreasing the concentration of IPTG caused growth inhibition after overnight incubation resulting in smaller and finally no colonies (on plates without IPTG) at 37°C. Table 3 shows the minimal concentration of IPTG at which still some growth was observed for the different conditional mutant strains. This concentration of IPTG was used in our experiments and defined as the condition causing limited induction. Most mutant strains indeed showed growth inhibition at lower concentration of IPTG. The mutant strain168IyqfY, the mutant for the putative gene encoding a homologue of the IspG protein (step 6) did not show any dependency on IPTG on plates (varying IPTG concentrations from 0 to 1 mM). Plates without IPTG incubated overnight resulted in the growth of normal colonies and the liquid cultures obtained growth curves comparable with wild type B. subtilis 168, independent of the concentration of IPTG. For some of the other mutant strains there was some growth of the bacterial cultures without IPTG as well, although severely impaired.
For all conditional knock out strains isoprene emission was measured at the concentration of IPTG with limited expression in comparison to 1 mM (full induction) and no IPTG (full repression).
Figure 3 represents the data for strain 168IyqiE, the conditional mutant strain for the first step in the biosynthetic pathway. Full induction of the dxs gene resulted in a normal growth curve of the bacterial culture and in an isoprene accumulation profile comparable to the profile of the wild type stain B. subtilis 168. Depletion of IPTG inhibited cell growth. The maximal concentration of isoprene accumulated in the flask and corrected for the amount of cells was 15 fold lower for the culture supplemented with 10 μM IPTG (8 μg/ m3 / OD600 nm) than for the culture with 1 mM IPTG (119 μg/ m3 / OD600 nm).
Table 3 shows the maximum concentration of isoprene accumulated in the flask calculated on the amount of cells present at that time point for all tested mutant strains. Mutant strains for putative genes yluB, yacM, yabH, yacN, and yqfP encoding enzymes for respectively, steps 2, 3, 4, 5, and 7 showed significant differences in isoprene emission, varying from a 5–28 fold decrease at limited expression levels in comparison to full induction.
The knock-out strain of ypgA, putatively encoding an isopentenyl diphosphate isomerase (step 8), is the only knock-out of a nonessential gene. The mutant strain showed a normal growth curve and did not show a significant difference in the emission of isoprene in comparison to the wild type strain B. subtilis 168.
We also tested the function of yqiD, encoding a homologue for farnesyl diphosphate synthase (FDP; 7), synthase IspA (step 9) of E. coli. Depletion of IPTG to 10 μM did not result in a significant change in the isoprene emission by the mutant strain, while it did show to be dependent on IPTG for growth.
Discussion
To identify genes involved in the biosynthesis of the highly volatile compound isoprene, an efficient online detection system for this terpenoid (precursor) in the air above bacterial cultures was developed. Since the B. subtilis strains 6051, 23059, and 23856 were known to emit isoprene (Kuzma et al. 1995), we used these strains to set up the detection system for B. subtilis 168 for which the genome has been sequenced (Kunst et al. 1997) and functional knock outs have been made. All four strains emitted isoprene in our experiment. The differences in the amounts of isoprene emitted corresponded with the levels as described before, where B. subtilis 6051 and B. subtilis 23059 emitted higher amounts than B. subtilis 23856 (Kuzma et al. 1995). Since B. subtilis 168 showed emission of isoprene, the use of the experimental set up to investigate the isoprene synthesis by the mutant strains was justified. From the profile of isoprene emission with a maximum at the transition of the growth curve from the logarithmic phase to the stationary phase, it was concluded that from this time point on maximum isoprene concentrations had been reached and that amounts sampled from the culture every 15 min exceeded the production of new isoprene. We could not detect isoprene production in three phases using standard incubation in a shaking flask as has been described for B. subtilis 6051 grown in a bioreactor.
The conditional knock out strain of the yqiE encoding DXS, the transketolase responsible for the first step in the biosynthesis of isoprenoids, appeared to be highly dependent on IPTG. The dramatic decrease in isoprene production at low concentration of IPTG supported the role of this gene in the biosynthetic pathway of isoprene. yqiE has been identified as dxs encoding 1-d-deoxyxylulose-5-phosphate in B. subtilis before (Harker and Bramley 1999; Hecht et al. 2001). However, the confirming results for this mutant strain proved that the experimental set up of our work was valid to investigate the function of the other putative genes. Apparently, the isoprene emission is severely decreased when genes in the pathway are expressed at lower level than in the wild type strain.
Growth at the end of the incubation after several hours, as shown for many of the liquid cultures under IPTG depletion or even without suppletion of IPTG, may be explained by the occurrence of reversions in the mutant strain (Zanen et al. 2006). Another explanation could be the presence of a not fully repressed promoter in the complementation strain. It has been reported that in the used vector pMUTIN3 the Pspac promoter can give some low expression even in the absence of IPTG (Vagner et al. 1998; Petit et al. 1998). For genes showing low levels of expression in the wild type strain it is easy to get a pseudo wild type level of expression in the mutant strain. This may be in particular the case for highly efficient enzymes in the biosynthetic pathway.
For the putative genes yluB, yacM, yabH, yacN, and yqfP, encoding homologues of the enzymatic steps 2, 3, 4, 5 and 7, the isoprene accumulation of the conditional knock outs by depletion of IPTG clearly proved the involvement of these genes in the biosynthetic pathway isoprene. B. subtilis 168IyqfY (step 6) did not depend on IPTG and therefore nothing can be concluded from the data obtained with this mutant strain other than that this enzyme may be highly efficient leading to complementation already obtained at the low expression levels caused by leaking activity of the Pspac promoter (Zanen et al. 2006).
The nonessential gene ypgA encodes a homologue protein for an isopentenyl diphosphate isomerase (Idi) (step 8). The B. subtilis gene ypgA encodes for a so-called isopentenyl diphosphate isomerase type II protein (Takagi et al. 2004), while the genome of E. coli harbours a gene for a different Idi type I enzyme (Hahn et al. 1999). Interestingly, all archaea contain the Idi type II and all eukaryotes contain Idi type I. In genomes of eubacteria both are found, but there are also genomes without any of the described genes (Boucher and Doolittle 2000). In contrast to all other genes investigated in this study, the gene ypgA encoding for the IdiII was shown to be a nonessential gene in B. subtilis (Takagi et al. 2004). We also observed that the knock-out mutant of the bacterial strain is viable and produces isoprene. The idiI gene of E. coli has also been shown to be nonessential. Deletion mutants of this gene were viable on minimal medium (Hahn et al. 1999). Our findings, that the isoprene emission is not influenced by the knock out of the ypgA gene, can be explained by the enzymatic mechanism of the IspH protein, step 7, before the isomerase. IDP and DMADP can be synthesized independently by the catalytic action of IspH. A hypothetical mechanism for this reaction has been described (Eisenreich et al. 2004). This supports the nonessential character of the gene. The presence of isoprene proves that the isomer DMADP is still synthesized in the mutant and that the cells do not depend on the isomerase for the production of DMADP from IDP only. The isomerase functions in the balance of IDP and DMADP as a salvage protein (Eisenreich et al. 2004).
The first enzymatic biosynthetic step downstream the formation of the precursors IDP and DMADP is catalyzed by IspA, farnesyl diphosphate synthase (step 9). Therefore the putative gene is not involved in the biosynthesis of isoprene itself. The observation that isoprene is still emitted while the expression of the gene was depleted is easily explained by its function in the pathway downstream of IDP biosynthesis. Although some accumulation of the precursors IDP and DMADP could be expected by blocking yqiD, no significant differences in the flux toward the isoprene emission were detected. Apparently the efflux of isoprene is not influenced by the changes in FDP synthesis downstream in the isoprenoid pathway. This can be regarded as a contradiction to the hypothesis that isoprene emission functions as an efflux of an overflow metabolite.
In this study the involvement of several B. subtilis genes in the MEP pathway to isoprene was established by studying isoprene emission of mutants. Five genes, essential for viability, yluB, yacM, yabH, yacN, and yqfP, were shown to be essential for the isoprene production as well. Where these genes were at first instance depicted based on homology of the encoding proteins, the results of this study proved the functional involvement of the genes in the biosynthesis of isoprenoids on a biochemical level. A knockout of the sixth candidate gene, yqfY, did not yield a reduction in isoprene emission, nor a growth retardation, which might be explained by a not fully repressed Pspac promoter present in vector pMUTIN3. Knock-outs of the two other genes investigated in this study, showing no growth retardation as well as no reduction in isoprene emission, behaved in accordance with expectation.
Although it remains unknown why isoprene is emitted by bacteria, the knowledge about the pathway to isoprene can be used for further investigations towards a better understanding of the metabolic flux to IDP and its physiological function. Next to that the IDP pool can be optimized by metabolic pathway engineering, creating a B. subtilis strain as an efficient Gram-positive host for the heterologous production of terpenoids, like the valuable plant-derived pharmaceuticals artemisinin and paclitaxel. The potential of using heterologous production organisms for the supply of terpenoids is already under investigation for E. coli and for several yeast strains (Martin et al. 2003; Lindahl et al. 2006; Ro et al. 2006; Dejong et al. 2006). For the purpose of metabolic pathway engineering it is interesting to search for a gene encoding isoprene synthase in B. subtilis as well. Evidence for an enzymatic bioconversion and the identification of this specific enzyme could give the possibility to block the efflux of isoprene thereby increasing the amount of isoprenoid available for the synthesis of terpenoids.
References
Boucher Y, Doolittle WF (2000) The role of lateral gene transfer in the evolution of isoprenoid biosynthesis pathways. Mol Microbiol 37:703–716
Cleveland CC, Yavitt JB (1998) Microbial consumption of atmospheric isoprene in a temperate forest soil. Appl Environ Microbiol 64:172–177
David S, Estramareix B, Fischer JC, Therisod M (1981) 1-Deoxy-d-threo-2-pentulose, the precursor of the 5-carbon chain of the thiazole of thiamine. J Am Chem Soc 103:7341–7342
David S, Estramareix B, Fischer JC, Therisod M (1982) The biosynthesis of thiamine—synthesis of [1,1,1,5-(H4)-H-2]-1-deoxy-d-threo-2-pentulose and incorporation of this sugar in biosynthesis of thiazole by Escherichia coli cells. J Chem Soc Perkin Trans 1:2131–2137
Dejong JM, Liu Y, Bollon AP, Long RM, Jennewein S, Williams D, Croteau RB (2006) Genetic engineering of taxol biosynthetic genes in Saccharomyces cerevisiae. Biotechnol Bioeng 93:212–224
Eisenreich W, Bacher A, Arigoni D, Rohdich F (2004) Biosynthesis of isoprenoids via the non-mevalonate pathway. Cell Mol Life Sci 61:1401–1426
Fall R, Copley SD (2000) Bacterial sources and sinks of isoprene, a reactive atmospheric hydrocarbon. Environ Microbiol 2:123–130
Hahn FM, Hurlburt AP, Poulter CD (1999) Escherichia coli open reading frame 696 is idi, a nonessential gene encoding isopentenyl diphosphate isomerase. J Bacteriol 181:4499–4504
Harker M, Bramley PM (1999) Expression of prokaryotic 1-deoxy-d-xylulose-5-phosphatases in Escherichia coli increases carotenoid and ubiquinone biosynthesis. FEBS Lett 448:115–119
Hecht S, Kis K, Eisenreich W, Amslinger S, Wungsintaweekul J, Herz S, Rohdich F, Bacher A (2001) Enzyme-assisted preparation of isotope-labeled 1-deoxy-d-xylulose 5-phosphate. J Org Chem 66:3948–3952
Hill RE, Sayer BG, Spenser ID (1989) Biosynthesis of vitamin B6—incorporation of d-1-deoxyxylulose. J Am Chem Soc 111:1916–1917
Kajiwara S, Fraser PD, Kondo K, Misawa N (1997) Expression of an exogenous isopentenyl diphosphate isomerase gene enhances isoprenoid biosynthesis in Escherichia coli. Biochem J 324 (Pt 2):421–426
Kaneda K, Kuzuyama T, Takagi M, Hayakawa Y, Seto H (2001) An unusual isopentenyl diphosphate isomerase found in the mevalonate pathway gene cluster from Streptomyces sp. strain CL190. Proc Natl Acad Sci U S A 98:932–937
Kim S, Keasling J (2001) Metabolic engineering of the nonmevalonate isopentenyl diphosphate synthesis pathway in Escherichia coli enhances lycopene production. Biotechnol Bioeng 72:408–415
Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, Boland F, Brignell SC, Bron S, Bunai K, Chapuis J, Christiansen LC, Danchin A, Debarbouille M, Dervyn E, Deuerling E, Devine K, Devine SK, Dreesen O, Errington J, Fillinger S, Foster SJ, Fujita Y, Galizzi A, Gardan R, Eschevins C, Fukushima T, Haga K, Harwood CR, Hecker M, Hosoya D, Hullo MF, Kakeshita H, Karamata D, Kasahara Y, Kawamura F, Koga K, Koski P, Kuwana R, Imamura D, Ishimaru M, Ishikawa S, Ishio I, Le Coq D, Masson A, Mauel C, Meima R, Mellado RP, Moir A, Moriya S, Nagakawa E, Nanamiya H, Nakai S, Nygaard P, Ogura M, Ohanan T, O’Reilly M, O’Rourke M, Pragai Z, Pooley HM, Rapoport G, Rawlins JP, Rivas LA, Rivolta C, Sadaie A, Sadaie Y, Sarvas M, Sato T, Saxild HH, Scanlan E, Schumann W, Seegers JF, Sekiguchi J, Sekowska A, Seror SJ, Simon M, Stragier P, Studer R, Takamatsu H, Tanaka T, Takeuchi M, Thomaides HB, Vagner V, van Dijl JM, Watabe K, Wipat A, Yamamoto H, Yamamoto M, Yamamoto Y, Yamane K, Yata K, Yoshida K, Yoshikawa H, Zuber U, Ogasawara N (2003) Essential Bacillus subtilis genes. Proc Natl Acad Sci USA 100:4678–4683
Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell SC, Bron S, Brouillet S, Bruschi CV, Caldwell B, Capuano V, Carter NM, Choi SK, Codani JJ, Connerton IF, Danchin A (1997) The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249–256
Kuzma J, Nemecek-Marshall M, Pollock WH, Fall R (1995) Bacteria produce the volatile hydrocarbon isoprene. Curr Microbiol 30:97–103
Kuzuyama T (2002) Mevalonate and nonmevalonate pathways for the biosynthesis of isoprene units. Biosci Biotechnol Biochem 66:1619–27
Lange BM, Rujan T, Martin W, Croteau R (2000) Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes. Proc Natl Acad Sci USA 97:13172–13177
Lindahl AL, Olsson ME, Mercke P, Tollbom O, Schelin J, Brodelius M, Brodelius PE (2006) Production of artemisinin precursor amorpha-4,11-diene by engineered Saccharomyces cerevisiae. Biotechnol Lett 28:571–580
Lois LM, Campos N, Putra SR, Danielsen K, Rohmer M, Boronat A (1998) Cloning and characterization of a gene from Escherichia coli encoding a transketolase-like enzyme that catalyzes the synthesis of d-1-deoxyxylulose 5-phosphate, a common precursor for isoprenoid, thiamin, and pyridoxol biosynthesis. Proc Natl Acad Sci USA 95:2105–2110
Loreto F, Delfine S (2000) Emission of isoprene from salt-stressed Eucalyptus globulus leaves. Plant Physiol 123:1605–1610
Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD (2003) Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol 21:796–802
Miller B, Oschinski C, Zimmer W (2001) First isolation of an isoprene synthase gene from poplar and successful expression of the gene in Escherichia coli. Planta 213:483–487
Petit MA, Dervyn E, Rose M, Entian KD, McGovern S, Ehrlich SD, Bruand C (1998) PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication. Mol Microbiol 29:261–273
Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, Chang MC, Withers ST, Shiba Y, Saprong R, Keasling JD (2006) Nature 440:940–943
Rohdich F, Kis K, Bacher A, Eisenreich W (2001) The non-mevalonate pathway of isoprenoids: genes, enzymes and intermediates. Curr Opin Chem Biol 5:535–540
Rohmer M (1999) The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Nat Prod Rep 16:565–574
Rohmer M, Knani M, Simonin P, Sutter B, Sahm H (1993) Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentenyl diphosphate. Biochem J 295 (Pt 2):517–524
Schnitzler JP, Zimmer I, Bachl A, Arend M, Fromm J, Fischbach RJ (2005) Biochemical properties of isoprene synthase in poplar (Populus x canescens). Planta 222:777–786
Seto H, Watanabe H, Furihata K (1996) Simultaneous operation of the mevalonate and non-mevalonate pathways in the biosynthesis of isopentenyl diphosphate in Streptomyces aeriouvifer. Tetrahedron Lett 37:7979–7982
Sivy TL, Shirk MC, Fall R (2002) Isoprene synthase activity parallels fluctuations of isoprene release during growth of Bacillus subtilis. Biochem Biophys Res Commun 294:71–75
Sprenger GA, Schorken U, Wiegert T, Grolle S, de Graaf AA, Taylor SV, Begley TP, Bringer-Meyer S, Sahm H (1997) Identification of a thiamin-dependent synthase in Escherichia coli required for the formation of the 1-deoxy-d-xylulose 5-phosphate precursor to isoprenoids, thiamin, and pyridoxol. Proc Natl Acad Sci U S A 94:12857–12862
Takagi M, Kaneda K, Shimizu T, Hayakawa Y, Seto H, Kuzuyama T (2004) Bacillus subtilis ypgA gene is fni, a nonessential gene encoding type 2 isopentenyl diphosphate isomerase. Biosci Biotechnol Biochem 68:132–137
Takahashi S, Kuzuyama T, Watanabe H, Seto H (1998) A 1-deoxy-d-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-d-erythritol 4-phosphate in an alternative nonmevalonate pathway for terpenoid biosynthesis. Proc Natl Acad Sci U S A 95:9879–9884
Vagner V, Dervyn E, Ehrlich SD (1998) A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144 (Pt 11):3097–3104
Wagner WP, Nemecek-Marshall M, Fall R (1999) Three distinct phases of isoprene formation during growth and sporulation of Bacillus subtilis. J Bacteriol 181:4700–4703
Wagner WP, Helmig D, Fall R (2000) Isoprene biosynthesis in Bacillus subtilis via the methylerythritol phosphate pathway. J Nat Prod 63:37–40
White RH (1978) Stable isotope studies on the biosynthesis of the thiazole moiety of thiamin in Escherichia coli. Biochemistry 17:3833–3840
Wilding EI, Brown JR, Bryant AP, Chalker AF, Holmes DJ, Ingraham KA, Iordanescu S, So CY, Rosenberg M, Gwynn MN (2000) Identification, evolution, and essentiality of the mevalonate pathway for isopentenyl diphosphate biosynthesis in gram-positive cocci. J Bacteriol 182:4319–4327
Yansura DG, Henner DJ (1984) Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc Natl Acad Sci U S A 81:439–443
Zanen G, Antelmann H, Meima R, Jongbloed JD, Kolkman M, Hecker M, van Dijl JM, Quax WJ (2006) Proteomic dissection of potential signal recognition particle dependence in protein secretion by Bacillus subtilis. Proteomics 6:3636–3648
Acknowledgements
The authors would like to express their gratitude to Dr. K. Kobayashi (Nara Institute of Science and Technology, Nara, Japan) for providing us with the Bacillus subtilis conditional knock-out strains for the essential genes and the BFAN project and Dr. S. Aymerich (Institute National de la Recherche Agronomique, Grignon, France) for contributing the ypgA knock-out mutant to the BFAN collection.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Julsing, M.K., Rijpkema, M., Woerdenbag, H.J. et al. Functional analysis of genes involved in the biosynthesis of isoprene in Bacillus subtilis . Appl Microbiol Biotechnol 75, 1377–1384 (2007). https://doi.org/10.1007/s00253-007-0953-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-007-0953-5