Abstract
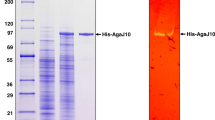
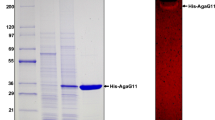
The agaA gene encoding β-agarase-a (AgaA) was cloned from the chromosomal DNA of a marine bacterium, Vibrio sp. strain PO-303. The nucleotide sequence of the agaA gene consists of 2,958 bp and encodes a protein of 985 amino acids with a molecular mass of 106,062 Da. The deduced enzyme protein contains a typical N-terminal signal peptide of 29 amino acid residues, followed by a 266 amino acid sequence that is homologous to catalytic module of family 16 glycoside hydrolases, a bacterial immunoglobulin group 2 (Big-2)-like domain of 52 amino acid residues, two carbohydrate-binding modules of family 6 separated from Big-2-like domain by nine times repeated GDDTDP amino acid sequence. AgaA is the first agarase that was identified to possess a Big-2-like domain. The recombinant AgaA (rAgaA) expressed in Escherichia coli exhibited maximal activity around 40°C and pH 7.5, with a specific activity of 16.4 units mg−1, a K m of 1.10 mg ml−1, and a V max of 22.5 μmol min−1 mg−1 for agarose. The rAgaA hydrolyzed neoagarohexaose, but did not act on neoagarotetraose and neoagarobiose.




Similar content being viewed by others
References
Allouch J, Jam M, Helbert W, Barbeyron T, Kloareg B, Henrissat B, Czjzek M (2003) The three-dimensional structures of two β-agarases. J Biol Chem 278:47171–47180
Aoki T, Araki T, Kitamikado M (1990) Purification and characterization of β-agarase from Vibrio sp. AP-2. Nippon Suisan Gakkaishi 56:825–830
Araki T, Hayakawa M, Zhang L, Karita S, Morishita T (1998a) Purification and characterization of agarases from a marine bacterium, Vibrio sp. PO-303. J Mar Biotechnol 6:260–265
Araki T, Zhang L, Morishita T (1998b) Optimization of parameters for isolation of protoplasts from Gracilaria verrucosa (Rhodophyta). J Mar Biotechnol 6:193–197
Bailey RW, Bourne EJ (1960) Colour reactions given by sugars and diphenylamine-aniline spray reagents on paper chromatograms. J Chromatogr 4:206–213
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Breslauer KJ, Frank R, Blocker H, Marky LA (1986) Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci USA 83:3746–3750
Coutinho PM, Henrissat B (1999) The modular structure of cellulases and other carbohydrate-active enzyme: an integrated database approach. In: Ohmiya K, Hayashi K, Sakka K, Kobayashi C, Karita S, Kimura T (eds) Genetics, biochemistry and ecology of cellulose degradation. Uni Publishers, Tokyo, Japan, pp 15–23
Dong J, Hashikawa S, Konishi T, Tamaru Y, Araki T (2006) Cloning of the novel gene encoding β-agarase C from a marine bacterium, Vibrio sp. strain PO-303, and characterization of the gene product. Appl Environ Microbiol 72:6399–6401
Ekborg NA, Taylor LE, Longmire AG, Henrissat B, Weiner RM, Hutcheson SW (2006) Genomic and proteomic analysis of the agarolytic system expressed by Saccharophagus degradans 2–40. Appl Environ Microbiol 72:3396–3405
Duckworth M, Yaphe W (1971) Structure of agar. I. Fractionation of a complex mixture of polysaccharides. Carbohydr Res 16:189–197
Ha JC, Kim GT, Kim SK, Oh TK, Yu JH, Kong IL (1997) β-Agarase from Pseudomonas sp. W7: purification of the recombinant enzyme from Escherichia coli and the effects of salt on its activity. Biotechnol Appl Biochem 26:1–6
Hahn M, Olsen O, Politz O, Borriss R, Heinemann U (1995) Crystal structure and site-directed mutagenesis of Bacillus macerans endo-1,3-1,4-β-glucanase. J Biol Chem 270:3081–3088
Henrissat B, Bairoch A (1996) Updating the sequence-based classification of glycosyl hydrolases. Biochem J 316:695–696
Henshaw J, Horne A, Bueren AL, Money VA, Bolam AN, Czjzek M, Ekborg NA, Weiner RM, Hutcheson SW, Davies GJ, Boraston AB, Gilbert HJ (2006) Family 6 carbohydrate binding modules in β-agarases display exquisite selectively for the non-reducing termini of agarose-chains. J Biol Chem 281:17099–17107
Jam M, Flament D, Allouch J, Potin P, Thion L, Kloareg B, Czjzek M, Helbert W, Michel G, Barbeyron T (2005) The endo-β-agarases AgaA and AgaB from the marine bacterium Zobelliagalactanivorans: two paralogue enzymes with different molecular organization and catalytic behaviors. Biochem J 385:703–713
Juncosa M, Pons J, Dot T, Querol E, Planas A (1994) Identification of active site carboxylic residues in Bacilluslicheniformis 1, 3-1, 4-β-d-glucan 4-glucanohydrolase by site-directed mutagenesis. J Biol Chem 269:14530–14535
Kang NY, Choi YL, Cho YS, Kim BK, Jeon BS, Cha JY, Kim CH, Lee YC (2003) Cloning, expression and characterization of a β-agarase gene from a marine bacterium, Pseudomonas sp. SK38. Biotechnol Lett 25:1165–1170
Kobayashi R, Takisada M, Suzuki T, Kirimura K, Usami S (1997) Neoagarobiose as a novel moisturizer with whitening effect. Biosci Biotechnol Biochem 61:162–163
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Luo Y, Frey EA, Pfuetzner RA, Creagh AL, Knoechel DG, Haynes CA, Finlay BB, Strynadka NC (2000) Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex. Nature 405(6790):1073–1077
McCarter JD, Withers SG (1994) Mechanisms of enzymatic glycoside hydrolysis. Curr Opin Struct Biol 4:885–892
Michel G, Chantalat L, Duee E, Barbeyron T, Henrissat B, Kloareg B, Dideberg O (2001) The κ-carrageenase of P. carrageenovora features a tunnel-shaped active site: a novel insight in the evolution of clan-B glycoside hydrolases. Structure 9:513–525
Ohta Y, Hatada Y, Nogi Y, Li Z, Ito S, Horikoshi K (2004a) Cloning, expression, and characterization of a glycoside hydrolase family 86 β-agarase from a deep-sea Microbulfifer-like isolate. Appl Microbiol Biotechnol 66:266–275
Ohta Y, Hatada Y, Miyazaki M, Nogi Y, Ito S, Horikoshi K (2004b) Purification and characterization of a novel α-agarase from a Thalassomonas sp. Curr Microbiol 50:212–216
Ohta Y, Hatada Y, Ito S, Horikoshi K (2005) High-level expression of a neoagarobiose-producing β-agarase gene from Agarivorans sp. JAMB-A11 in Bacillus subtilis and enzymatic properties of the recombinant enzyme. Biotechnol Appl Biochem 41:183–191
Potin P, Richard C, Rochas C, Kloareg B (1993) Purification and characterization of the α-agarase from Alteromonas agarlyticus (Cataldi) comb. nov., strain GJ1B. Eur J Biochem 214:599–607
Rosenberg M, Court D (1979) Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet 13:319–353
Saito H, Miura K (1963) Preparation of transforming deoxyribonucleic acid by phenol-treatment. Biochem Biophys Acta 72:619–629
Shine J, Dalgarno L (1975) Determinant of cistron specificity in bacterial ribosomes. Nature 254:34–38
Somogyi M (1952) Notes on sugar determination. J Biol Chem 195:19–23
Sugano Y, Matsumoto T, Kodama H, Noma M (1993) Cloning and sequencing of agaA, a unique agarase 0107 gene from a marine bacterium, Vibrio sp. strain JT0107. Appl Environ Microbiol 59:3750–3756
Sugano Y, Matsumoto T, Noma M (1994) Sequence analysis of the agaB gene encoding a new β-agarase from Vibrio sp. strain JT0107. Biochem Biophys Acta 1218:105–108
Sumimoto N, Nakano S, Katoh M, Matsumura A, Nakamura H, Ohmichi T, Yoneyama M, Sasaki M (1995) Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry 34:11211–11216
Tompson J, Higgins D, Gibson T (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Vera J, Alvarez R, Murano E, Slebe JC, Leon O (1998) Identification of a marine agarolytic Pseudoalteromonas isolate and characterization of its extracellular agarase. Appl Environ Microbiol 64:4378–4383
Yoshizawa Y, Ametani A, Tsunehiro J, Nomura K, Itoh M, Fukui F, Kaminogawa S (1995) Macrophage stimulation activity of the polysaccharide fraction from a marine alga (Porphyra yezoensis): structure-function relationships and improved solubility. Biosci Biotechnol Biochem 59:1933–1937
Acknowledgment
This study was supported by a grant program of the Agriculture, Forestry and Fisheries Research Council of Japan (Research Project for Utilizing Advanced Technologies in Agriculture, Forestry, and Fisheries no. 1681, 2004–2006).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dong, J., Tamaru, Y. & Araki, T. A unique β-agarase, AgaA, from a marine bacterium, Vibrio sp. strain PO-303. Appl Microbiol Biotechnol 74, 1248–1255 (2007). https://doi.org/10.1007/s00253-006-0781-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-006-0781-z