Abstract
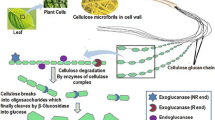
cDNAs encoding two glycoside hydrolase family 1 β-glucosidases (BGL1A and BGL1B) were cloned from the basidiomycete Phanerochaete chrysosporium, and the substrate specificities of the recombinant enzymes and the expression patterns of the two genes were investigated in relation to cellobiose metabolism by the fungus. The cDNA sequences contained open reading frames of 1,389 base pairs (bp) (bgl1A) and 1,623 bp (bgl1B), encoding 462 and 530 amino acids, respectively. Although high sequence identity (65%) was observed between the deduced amino acid sequences of the two enzymes, an apparent difference was observed at the C-terminal region: BGL1B has a 63-amino acid extension, which has no similarity with any known protein. Both recombinant enzymes expressed in Escherichia coli showed hydrolytic activity towards several β-glycosidic compounds. However, the substrate recognition patterns of the two enzymes were quite different from each other. In particular, cellobiose was hydrolyzed more effectively by BGL1B than by BGL1A. The expression of the two genes in the fungus was monitored by reverse transcription-PCR, which showed that bgl1A was expressed constitutively in both glucose- and cellobiose-containing culture, whereas bgl1B was expressed in cellobiose culture but was repressed in glucose culture, possibly because of carbon catabolite repression. We conclude that BGL1B contributes to cellobiose metabolism during cellulose degradation by P. chrysosporium.





Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Ayers AR, Ayers SB, Eriksson KE (1978) Cellobiose oxidase, purification and partial characterization of a hemoprotein from Sporotrichum pulverulentum. Eur J Biochem 90:171–181
Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: signalP 3.0. J Mol Biol 340:783–795
Broda P, Birch PR, Brooks PR, Sims PF (1995) PCR-mediated analysis of lignocellulolytic gene transcription by Phanerochaete chrysosporium: substrate-dependent differential expression within gene families. Appl Environ Microbiol 61:2358–2364
Eriksson KE (1978) Enzyme mechanisms involved in cellulose hydrolysis by rot fungus Sporotrichum pulverulentum. Biotechnol Bioeng 20:317–332
Eriksson KE, Pettersson B, Westermark U (1974) Oxidation: an important enzyme reaction in fungal degradation of cellulose. FEBS Lett 49:282–285
Habu N, Igarashi K, Samejima M, Pettersson B, Eriksson KE (1997) Enhanced production of cellobiose dehydrogenase in cultures of Phanerochaete chrysosporium supplemented with bovine calf serum. Biotechnol Appl Biochem 26:97–102
Henrissat B (1991) A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 280:309–316
Henrissat B, Bairoch A (1993) New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 293:781–788
Henrissat B, Bairoch A (1996) Updating the sequence-based classification of glycosyl hydrolases. Biochem J 316:695–696
Igarashi K, Samejima M, Saburi Y, Habu N, Eriksson KE (1997) Localization of cellobiose dehydrogenase in cellulose-grown cultures of Phanerochaete chrysosporium. Fungal Genet Biol 21:214–222
Igarashi K, Tani T, Kawai R, Samejima M (2003) Family 3 β-glucosidase from cellulose-degrading culture of the white-rot fungus Phanerochaete chrysosporium is a glucan 1,3-β-glucosidase. J Biosci Bioeng 95:572–576
Johnsrud SC, Eriksson KE (1985) Cross-breeding of selected and mutated homokaryotic strains of Phanerochaete chrysosporium K-3—new cellulase deficient strains with increased ability to degrade lignin. Appl Microbiol Biotechnol 21:320–327
Kawai R, Yoshida M, Tani T, Igarashi K, Ohira T, Nagasawa H, Samejima M (2003) Production and characterization of recombinant Phanerochaete chrysosporium β-glucosidase in the methylotrophic yeast Pichia pastoris. Biosci Biotechnol Biochem 67:1–7
Kawai R, Igarashi K, Kitaoka M, Ishii T, Samejima M (2004) Kinetics of substrate transglycosylation by glycoside hydrolase family 3 glucan (1->3)-β-glucosidase from the white-rot fungus Phanerochaete chrysosporium. Carbohydr Res 339:2851–2857
Kawai R, Igarashi K, Yoshida M, Kitaoka M, Samejima M (2005) Hydrolysis of β-1,3/1,6-glucan by glycoside hydrolase family 16 endo-1,3(4)-β-glucanase from the basidiomycete Phanerochaete chrysosporium. Appl Microbiol Biotechnol (in press) https://doi.org/10.1007/s00253-005-0214-4
Kremer SM, Wood PM (1992) Evidence that cellobiose oxidase from Phanerochaete chrysosporium is primarily an Fe(III) reductase—kinetic comparison with neutrophil NADPH oxidase and yeast flavocytochrome b 2. Eur J Biochem 205:133–138
Li B, Renganathan V (1998) Gene cloning and characterization of a novel cellulose-binding β-glucosidase from Phanerochaete chrysosporium. Appl Environ Microbiol 64:2748–2754
Lymar ES, Li B, Renganathan V (1995) Purification and characterization of a cellulose-binding β-glucosidase from cellulose-degrading cultures of Phanerochaete chrysosporium. Appl Environ Microbiol 61:2976–2980
Martinez D, Larrondo LF, Putnam N, Gelpke MD, Huang K, Chapman J, Helfenbein KG, Ramaiya P, Detter JC, Larimer F, Coutinho PM, Henrissat B, Berka R, Cullen D, Rokhsar D (2004) Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol 22:695–700
Mchale A, Coughlan MP (1981a) The cellulolytic system of Talaromyces emersonii—identification of the various components produced during growth on cellulosic media. Biochim Biophys Acta 662:145–151
Mchale A, Coughlan MP (1981b) The cellulolytic system of Talaromyces emersonii—purification and characterization of the extracellular and intracellular β-glucosidases. Biochim Biophys Acta 662:152–159
Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10:1–6
Peralta RM, Terenzi HF, Jorge JA (1990) β-D-Glycosidase activities of Humicola grisea: biochemical and kinetic characterization of a multifunctional enzyme. Biochim Biophys Acta 1033:243–249
Saloheimo M, Kuja-Panula J, Ylosmaki E, Ward M, Penttila M (2002) Enzymatic properties and intracellular localization of the novel Trichoderma reesei β-glucosidase BGLII (cel1A). Appl Environ Microbiol 68:4546–4553
Smith MH, Gold MH (1979) Phanerochaete chrysosporium β-glucosidases: induction, cellular localization, and physical characterization. Appl Environ Microbiol 37:938–942
Takashima S, Nakamura A, Masaki H, Uozumi T (1996) Purification and characterization of cellulases from Humicola grisea. Biosci Biotechnol Biochem 60:77–82
Takashima S, Nakamura A, Hidaka M, Masaki H, Uozumi T (1999) Molecular cloning and expression of the novel fungal β-glucosidase genes from Humicola grisea and Trichoderma reesei. J Biochem (Tokyo) 125:728–736
Tempelaars CA, Birch PR, Sims PF, Broda P (1994) Isolation, characterization, and analysis of the expression of the cbhII gene of Phanerochaete chrysosporium. Appl Environ Microbiol 60:4387–4393
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Westermark U, Eriksson KE (1974) Cellobiose: quinone oxidoreductase, a new wood-degrading enzyme from white-rot fungi. Acta Chem Scand B 28:209–214
Yoshida M, Ohira T, Igarashi K, Nagasawa H, Aida K, Hallberg BM, Divne C, Nishino T, Samejima M (2001) Production and characterization of recombinant Phanerochaete chrysosporium cellobiose dehydrogenase in the methylotrophic yeast Pichia pastoris. Biosci Biotechnol Biochem 65:2050–2057
Yoshida M, Igarashi K, Kawai R, Aida K, Samejima M (2004) Differential transcription of β-glucosidase and cellobiose dehydrogenase genes in cellulose degradation by the basidiomycete Phanerochaete chrysosporium. FEMS Microbiol Lett 235:177–182
Acknowledgements
The authors are grateful to Dr. Shinya Fushinobu (Department of Biotechnology, University of Tokyo) for valuable discussions about the kinetics of GH family 1 BGL. This research was supported by a Grant-in-Aid for Scientific Research to M. Samejima (number 17380102) from the Japanese Ministry of Education, Culture, Sports and Technology.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Tsukada, T., Igarashi, K., Yoshida, M. et al. Molecular cloning and characterization of two intracellular β-glucosidases belonging to glycoside hydrolase family 1 from the basidiomycete Phanerochaete chrysosporium . Appl Microbiol Biotechnol 73, 807–814 (2006). https://doi.org/10.1007/s00253-006-0526-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-006-0526-z