Abstract
Antimicrobial peptides or bacteriocins are excellent candidates for alternative antimicrobials, but high manufacturing costs limit their applications. Recombinant gene expression offers the potential to produce these peptides more cost-effectively at a larger scale. Saccharomyces cerevisiae is a popular host for recombinant protein production, but with limited success reported on antimicrobial peptides. Individual recombinant S. cerevisiae strains were constructed to secrete two class IIa bacteriocins, plantaricin 423 (PlaX) and mundticin ST4SA (MunX). The native and codon-optimised variants of the plaA and munST4SA genes were cloned into episomal expression vectors containing either the S. cerevisiae alpha mating factor (MFα1) or the Trichoderma reesei xylanase 2 (XYNSEC) secretion signal sequences. The recombinant peptides retained their activity and stability, with the MFα1 secretion signal superior to the XYNSEC secretion signal for both bacteriocins. An eight-fold increase in activity against Listeria monocytogenes was observed for MunX after codon optimisation, but not for PlaX-producing strains. After HPLC-purification, the codon-optimised genes yielded 20.9 mg/L of MunX and 18.4 mg/L of PlaX, which displayed minimum inhibitory concentrations (MICs) of 108.52 nM and 1.18 µM, respectively, against L. monocytogenes. The yields represent a marked improvement relative to an Escherichia coli expression system previously reported for PlaX and MunX. The results demonstrated that S. cerevisiae is a promising host for recombinant bacteriocin production that requires a simple purification process, but the efficacy is sensitive to codon usage and secretion signals.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
With the increase in antibiotic resistance among bacterial strains, there is an urgent need for alternative antimicrobial agents. Some of the most potent and specific antimicrobials are antimicrobial peptides (AMPs), which are small, cationic peptides (10 to 100 amino acid residues) with an amphipathic structure and a net positive charge [1]. Bacteriocins of gram-positive bacteria are classified into three groups based on their size, structure and modifications [2]. Class IIa bacteriocins, also known as pediocin-like bacteriocins (named after pediocin PA-1, the first bacteriocin characterised in this group), are small (usually less than 10 kDa) and heat-stable, with post-translational modifications that are limited to disulphide bond formation [3, 4]. These peptides contain a conserved N-terminal YGNGV motif (pediocin box) that serves as a recognition sequence for the membrane-bound protein receptor (protein IIC (MptC)) of mannose phosphotransferase (Man-PTS) in target cells [5, 6]. Interaction of the AMP with the IIC docking molecule disrupts the Man-PTS system by preventing the transport of sugars required for growth, causing cell membrane permeabilisation and cell death [4, 7].
Class IIa bacteriocins have been investigated mainly for their application as natural food preservatives, such as antimicrobial packaging and coating fresh produce and meat products [8,9,10]. Peptides from this subclass mainly target related lactic acid bacteria and different species of Staphylococcus and Listeria. This includes Listeria monocytogenes, the deadliest bacterial source of food poisoning, with 30% of infections in high-risk individuals being fatal [2]. However, there is a growing interest in their application as therapeutic antimicrobial agents as they are highly potent at low concentrations, and their narrow specificity reduces their impact on commensal microbiota [2, 11]. Class IIa bacteriocins are also active against vancomycin-resistant enterococci and Staphylococcus aureus, among others [12, 13].
The probiotic lactic acid bacteria, Lactiplantibacillus plantarum 423 (previously Lactobacillus plantarum 423) and Enterococcus mundtii ST4SA, produce the class IIa bacteriocins plantaricin 423 (PlaX) and mundticin ST4SA (MunX), respectively. These AMPs are active against various foodborne pathogens, including Bacillus cereus, Clostridium sporogenes, Enterococcus faecalis, L. monocytogenes and S. aureus [14, 15]. Van Zyl et al. [16] demonstrated the importance of these bacteriocins as probiotic anti-infective mediators in L. plantarum 423 and E. mundtii ST4SA to inhibit the growth of L. monocytogenes in the gastrointestinal tract of infected mice.
Production of class IIa bacteriocins from their natural producer strains requires the expression of several genes clustered into one or more operons [17]. These operons contain genes that encode a pre-peptide, immunity protein, ATP-binding cassette (ABC) transporter, accessory proteins for the extracellular translocation of the peptide, accessory disulphide modification proteins and regulatory proteins [18]. The complex regulatory machinery can limit or cause inconsistent peptide production from the native host [19, 20]. AMP production by the native host can also be expensive as rich and complex medium is required for improved bacterial growth and peptide production [21]. Moreover, several purification steps are required to purify the peptides from complex growth medium, further inflating the cost of large-scale AMP production.
Recombinant gene expression could allow the more cost-effective and consistent production of AMPs with the potential to scale up the production process. Escherichia coli is the most widely used host for producing foreign proteins, but this is hampered by the bacterial host’s sensitivity towards AMPs [22]. Furthermore, E. coli does not naturally secrete proteins into the extracellular medium, which can lead to insoluble inclusion bodies and the accumulation of non-functional proteins due to misfolding and a high rate of protein degradation by intracellular proteases [23]. Different strategies have been investigated to overcome this, including signal peptides to facilitate secretion and fusion proteins to mask the activity of AMPs, thus reducing AMP toxicity to the producer strain and preventing proteolytic cleavage [24]. Recombinant plantaricin 423 and mundticin ST4SA were expressed in E. coli by fusing the mature peptides to a His-tagged green fluorescent protein (GFP) [25]. The fusion proteins reduced the bacteriocins’ toxicity, but additional purification steps increased AMP production costs and reduced the yields.
The yeast Saccharomyces cerevisiae offers a promising alternative to produce class IIa bacteriocins as the cells are not sensitive to AMPs. It can also perform post-translational modification of peptides, such as disulphide bond formation catalysed by two enzymes in the endoplasmic reticulum (ER), namely ER oxidoreductin and protein disulphide isomerase (PDI) [26]. The yeast does not require complex growth media and can efficiently secrete peptides extracellularly, thus increasing the protein titers and reducing the need for additional purification steps [27]. Strains of S. cerevisiae are used extensively to produce recombinant proteins and enzymes [28, 29], and a few studies reported the expression of bacteriocins in S. cerevisiae [30,31,32,33]. Schoeman et al. [30] constructed a bactericidal S. cerevisiae Y294 strain that expressed the native pediocin PA-1, while Van Reenen et al. [31] heterologously expressed the native plantaricin 423 in S. cerevisiae L5366h. However, the expression of both bacteriocins was driven by the inducible yeast alcohol dehydrogenase I promoter (ADH1P) and terminator (ADH1T), which yielded low titres.
Different strategies could be employed to improve the heterologous expression of these peptides in S. cerevisiae. One approach is to implement codon optimisation, which alters the codon usage pattern using synonymous codons to match the codon usage bias of the native host organism [34]. A secretion signal that targets the AMP to the extracellular matrix would simplify purification, avoid peptide degradation by intracellular proteases and reduce peptide misfolding as chaperone proteins direct the peptides for proper folding and secretion [35]. A strong constitutive promoter and terminator could ensure gene transcription throughout the cultivation process, resulting in higher levels of recombinant peptide [36].
This study aimed to develop an optimised expression system for producing class IIa bacteriocins in S. cerevisiae, using the L. plantarum 423 and E. mundtii ST4SA bacteriocins as benchmarks. Codon-optimised and native variants of plaA and munST4SA were expressed in S. cerevisiae Y294 under the control of the constitutive ENO1 promoter and terminator. Furthermore, the native S. cerevisiae mating factor-alpha secretion signal (MFα1) [37] and the Trichoderma reesei xylanase 2 secretion signal (XYNSEC) [38] were evaluated for secretion of the peptides to the extracellular medium.
Materials and Methods
Strains and Plasmids
The yeast and bacterial strains used and constructed in this study are listed in Table 1, and the plasmids in Table 2.
Media and Cultivation Conditions
All media components and reagents were sourced from Merck (Darmstadt, Germany) unless stated otherwise. The E. coli DH5α strain (Takara Bio Inc., Japan) was used for plasmid propagation; the transformants were maintained and selected for on Luria Bertani (LB) agar containing ampicillin (100 µg/mL) for selective pressure. Transformants were routinely cultured at 37 °C in Terrific Broth (12 g/L tryptone, 24 g/L yeast extract, 4 m/L glycerol, 0.1 M potassium phosphate buffer; pH 7.0) containing 100 µg/mL ampicillin [39]. The E. coli BL21 (DE3) PlantEx and MunEx strains, expressing the native plaA and munST4SA genes, respectively [25], were maintained on Brain Heart Infusion (BHI) agar supplemented with 50 µg/mL kanamycin.
The S. cerevisiae Y294 strain served as the host for recombinant gene expression and was maintained on YPD agar (10 g/L yeast extract, 20 g/L peptone, 20 g/L glucose, 20 g/L agar) and routinely cultured in YPD broth at 30 °C. Recombinant strains were selected for and maintained on SC−URA agar (6.7 g/L yeast nitrogen base without amino acids (BD Diagnostic Systems, Maryland, USA), 20 g/L glucose, 1.5 g/L synthetic drop-out medium supplements without uracil (Sigma-Aldrich, Steinheim, Germany) and 20 g/L agar; pH 6.0). All S. cerevisiae strains were aerobically cultivated on a rotary shaker (200 rpm) at 30 °C in 125 mL Erlenmeyer flasks containing 20 mL double-strength SC−URA broth (2 × SC−URA) as outlined in [41]. Unless stated otherwise, all growth media were inoculated at a final cell count of 1 × 106 CFU/mL.
Listeria monocytogenes EDG-e was used as the indicator strain for all antimicrobial activity assays. The strain was maintained on BHI agar supplemented with 7.5 µg/mL chloramphenicol and incubated at 37 °C.
Construction of Recombinant Strains
Gene Design and Synthesis
The active bacteriocin-coding sequences for the native plaA and munST4SA genes were obtained from plasmids pRSF-GFP-PlaX and pRSF-GFP-MunX [25]. The DNA sequences of the plaA and munST4SA genes encoding the mature bacteriocins (excluding the leader peptides) were codon-optimised by GenScript® (Piscataway, New Jersey, USA) using the OptimumGene™ algorithm for expression in S. cerevisiae. The resulting plasmids, pUC57-MFα1-PlaX_Opt and pUC57-MFα1-MunX_Opt, contained the native S. cerevisiae MFα1 secretion signal and the Kex2 and two Ste13 sites for protease cleavage at the N-termini of the peptides.
DNA Manipulations
DNA manipulations and E. coli transformations were performed using standard protocols [39]. The various native and codon-optimised nucleotide sequences of the peptides were amplified from plasmid DNA with PCR using primers designed for yeast-mediated ligation (YML) (Online Resource 1). The PCR amplifications were performed with the Gene Amp® PCR System 2400 Thermal Cycler (Perkin Elmer, Waltham, Massachusetts, United States) and TaKaRa Ex Taq™ (Takara Bio Inc.) as per the manufacturer’s recommendations. The PCR products were separated with 2% (w/v) agarose gel electrophoresis and isolated using the Zymoclean™ Gel DNA Recovery kit (Zymo Research, Irvine, California, USA).
The expression cassettes were constructed using yBBH1 and yBBH4 as plasmids backbones. Plasmid yBBH1 contains the ENO1 cassette, whereas yBBH4 has an additional XYNSEC secretion signal of the Trichoderma reesei xylanase 2 gene [38] cloned immediately downstream of the ENO1 promoter (Table 2; Online Resource 2). yBBH4 served as an expression vector for both the native and codon-optimised bacteriocins, whereas only the codon-optimised bacteriocins fused to the MFα1 secretion signal were cloned into yBBH1. NruI-linearised yBBH4 and EcoRI, XhoI-digested yBBH1 (restriction enzymes sourced from Inqaba Biotec, Pretoria, South Africa) were separated on a 0.8% (w/v) agarose gel and recovered using the Zymoclean™ Gel DNA Recovery kit (Zymo Research).
The PCR products were co-transformed with the linearised plasmids into electrocompetent S. cerevisiae Y294 cells [42] to construct in-frame fusions via YML with the enolase 1 (ENO1) promoter and terminator (Online Resource 2). Positive transformants were identified with colony-PCR using gene-specific primers (Online Resource 1) and Sanger sequencing (ABI PRISM™ 3100 Genetic Analyser, Central Analytical Facility, Stellenbosch University) of the plasmid DNA isolated from the yeast strains using the High Pure Plasmid Isolation kit (Roche, Mannheim, Germany).
The MFα1 gene was amplified from the MFα1-PlaX_Opt synthetic gene with the pMR-MFα1 primer set (Online Resource 1) and transformed together with linearised yBBH1 (EcoRI + XhoI) into electrocompetent S. cerevisiae Y294 cells to yield yBBH1-MFα1 (hereafter called pMR, Online Resource 2). The active bacteriocin-coding plaA and munST4SA genes were amplified from the pRSF-GFP-PlaX and pRSF-GFP-MunX plasmids using the MR-PlaX and MR-MunX primer sets (Online Resource 1), respectively, and transformed together with XhoI-linearised pMR to yield plasmids pMR-PlaX and pMR-MunX via YML (Online Resource 2). The gene cassettes are summarised in Online Resource 3, and the nucleotide and amino acid sequences of the secretion signals and various bacteriocins are listed in Online Resource 4.
Screening for Antimicrobial Activity
The recombinant yeast strains were screened for antimicrobial activity against L. monocytogenes EDG-e using agar-overlay and agar well-diffusion assays [30, 43, 44]. The S. cerevisiae strains transformed with yBBH1, yBBH4 or pMR (no AMP inserts) served as negative controls. Assays were performed in triplicate, and the average and standard deviation of the diameter of the inhibition zones were determined.
Agar-Overlay Assay
Preliminary screening for antimicrobial activity of the various recombinant yeast strains was conducted using the agar-overlay method [30], with a few modifications. Briefly, recombinant strains were grown overnight at 30 °C in test tubes containing 10 mL of SC−URA broth, 2 µL of each culture were spotted onto SC−URA plates and incubated for 72 h at 30 °C. The plates were overlaid with BHI 0.7% (w/v) agar seeded with a 1% (v/v) overnight culture of L. monocytogenes EDG-e. After incubation at 30 °C for 18 h, the plates were examined for inhibition zones.
Agar Well-Diffusion Assay
Erlenmeyer flasks (125 mL) containing 20 mL 2 × SC−URA broth were inoculated with 1 × 106 cells/mL of the respective recombinant S. cerevisiae strain and grown aerobically on a rotary shaker at 200 rpm for 72 h at 30 °C. The cell-free supernatant (CFS) was harvested (1500 × g, 5 min) and filtered through 0.2 µm pore-size low-protein binding non-pyrogenic membranes (syringe filters, Pall Life Sciences, New York, USA). The antimicrobial activity of the CFS was determined using the agar well-diffusion assay [43] by spotting 100 µL of each sample in 6-mm wells that were cut into the surface of BHI 1% (w/v) agar seeded with a 1% (v/v) overnight culture of L. monocytogenes EDG-e. All plates were incubated at 37 °C for 18 h and examined for inhibition zones. In addition, the proteinaceous nature of the active supernatant was verified by treating the samples with 10 µg/mL trypsin (in 10 mM TRIS buffer, pH 8) for 1 h at 37 °C, and then determining activity against L. monocytogenes EDG-e as described above.
Bacteriocin Activity Assays
Following the screening for antimicrobial activity, the bacteriocin activity of the recombinant yeast strains was evaluated over 72 h. The strains were cultivated as described above, and 1-mL aliquots of the respective strains were sampled every 24 h. Samples were centrifuged (1500 × g, 5 min), and the supernatant was filtered as described above and assessed for antimicrobial activity [45]. Briefly, a serial two-fold dilution of each sample was made in sterile 1 × phosphate-buffered saline (PBS, pH 7.4), and 100 µL of each diluted sample was tested for activity using the agar well-diffusion assay. The antimicrobial activity of the recombinant strain was expressed as arbitrary units per millilitre (AU/mL), corresponding to the reciprocal of the highest dilution that inhibited the indicator strain [46]. To determine the dry cell weight (DCW) after 72 h of growth, the cells in 1 mL of the supernatant were harvested by centrifugation (1500 × g for 5 min), dried overnight at 60 °C and weighed. Bacteriocin activity assays were performed in three biological repeats, and the average and standard deviations were determined.
Peptide Analysis Using Tricine-SDS-PAGE
The CFS from the recombinant strains were analysed using tricine SDS-PAGE [47] to confirm the production of the recombinant peptides. Before analysis, CFS was lyophilised for 3 days and dissolved in sterile 1X PBS to achieve a 20-fold concentration. Tricine SDS-PAGE analyses were performed in duplicate using the ultra-low range molecular weight marker (Sigma-Aldrich). One gel was subjected to Coomassie blue staining [47], followed by silver staining [48] to improve the visualisation of protein bands. The other gel was fixed for 20 min in a 25% (v/v) isopropanol, 10% (v/v) acetic acid fixing solution and rinsed thrice for 15 min with sterile Milli-Q water. The gel was then cast in a BHI 0.8% (w/v) agar bilayer (supplemented with 7.5 µg/mL chloramphenicol) seeded with an overnight culture of L. monocytogenes EDG-e and incubated overnight at 37 °C to assess antimicrobial activity [44].
Stability Tests
The antimicrobial activity against L. monocytogenes EDG-e was used to assess the stability of the recombinant peptides under different pH and temperature conditions, as outlined in Meng et al. [49]. Briefly, the recombinant strains were cultivated in 2 × SC−URA broth at 30 °C for 72 h and the CFSs harvested (1500 × g for 5 min). The supernatant was filtered as described elsewhere and exposed to a range of pH 2.0 to pH 10.0 (adjusted with either 1 N NaOH or 10 N HCl) and temperatures (4 °C, 30 °C, 37 °C, 60 °C, 80 °C and 100 °C), respectively, for 1 h. After treatment, the pH-treated samples were re-adjusted to the initial pH values, and the temperature-treated samples were allowed to return to room temperature. The antimicrobial activity of the treated samples was determined using the agar well-diffusion assay described above, and the diameter of the inhibition zones was measured. The S. cerevisiae Y294[MR] strain and untreated samples were included as controls. Stability tests were performed in duplicate, and an average value and standard deviation were determined for the inhibition zones.
Scanning Electron Microscopy (SEM)
Scanning electron microscopy was performed as per Reichhardt et al. [50], but with a few modifications. Listeria monocytogenes EDG-e was cultured to mid-logarithmic phase; the cells were harvested (2400 × g, 10 min), washed three times with 1X PBS and resuspended to an A600 value of 0.2. The bacterial cells were treated with equal volumes of 20-fold concentrated CFS of Y294[MFα1-PlaX_Opt], Y294[MFα1-MunX_Opt] and the Y294[MR] negative control, respectively, at 37 °C for 18 h. The cells were harvested as described elsewhere and washed thrice with 1X PBS. Samples were fixed overnight in 2.5% (v/v) glutaraldehyde with 4% (v/v) paraformaldehyde (PFA) in 0.1 M sodium-cacodylate buffer (pH 7.3) at 4 °C, washed twice in 0.1 M sodium-cacodylate and thrice in sterile Milli-Q water. The cell pellets were dehydrated in a graded ethanol series (30%, 50%, 70%, and 90%) for 15 min each and washed twice with 100% ethanol for 15 min at room temperature. The dried samples were treated with 50% (v/v) hexamethyldisilazane (HMDS) in ethanol for 15 min, followed by 100% HMDS for 15 min and allowed to air-dry overnight. Dried samples were mounted on 10 mm aluminium scanning electron microscopy (SEM) stubs with carbon tape and were coated with gold (10 nm) using a Leica EM ACE200 sputter-coater (Leica Microsystems, Germany) to enhance conductivity. The SEM imaging was completed using a Zeiss Merlin Field Emission SEM (Carl Zeiss Microscopy, Germany) operated at a 2–3 kV accelerating voltage, 89–100 pA beam current and using InLens Secondary Electron (SE) and SE2 detection.
Peptide Purification
The Y294[MFα1-PlaX_Opt] and Y294[MFα1-MunX_Opt] strains were cultivated as described above, and the CFS was harvested and filter-sterilised. The peptides (henceforth referred to as PlaX_Opt and MunX_Opt) were precipitated from the respective supernatants by adding acetone and trichloracetic acid (TCA) in 1:8:1 solvent ratio for 1 h at − 20 °C. Each mixture was centrifuged at 18,000 × g for 15 min at 4 °C, and the supernatant was discarded. Pellets were washed thrice with ice-cold acetone and dried at room temperature. The dried pellets were then dissolved in 10% (v/v) acetonitrile and tested for activity against L. monocytogenes EDG-e using the agar well-diffusion assay.
PlaX_Opt and MunX_Opt were partially purified from the respective active supernatant extractions using reverse-phase High-Performance Liquid Chromatography (HPLC) with the Agilent 1260 Infinity HPLC system and the ZORBAX 300SB-C8 column (4.6 × 150 mm, 5 µm particle size) (Agilent, California, United States). Sample separation was achieved using linear gradient elution from 10% solvent B to 60% solvent B over 25 min (solvent B: acetonitrile + 0.1% (v/v) trifluoroacetic acid (TFA) against solvent A: analytically pure water + 0.1% (v/v) TFA). Elution profiles were monitored at 230 and 214 nm and collected in 1 mL fractions. Acetonitrile was removed from the fractions under vacuum using a SpeedyVac vacuum concentrator (Savant), and the samples were tested against L. monocytogenes EDG-e using the agar well-diffusion assay [25].
Yield Estimation
To calculate the yield of PlaX_Opt and MunX_Opt, the recombinant strains were cultured in 100 mL of 2 × SC−URA broth, and the peptides were harvested and purified as described elsewhere. The HPLC-purified fractions that showed anti-listerial activity were combined, lyophilised and analytically weighed using an XP26 Excellence Plus Micro Balance (Mettler Toledo, Columbus, Ohio, USA). The yields were determined as the mean of three biological repeats in triplicate technical repeats. The purity of the HPLC-purified peptides was confirmed with tricine SDS-PAGE analysis and agar overlays. A concentration range (10 µg–312.5 ng) of bovine serum albumin (BSA) was included as a control for densitometry.
Liquid Chromatography and Tandem Mass Spectrometry Analysis (LC–MS/MS)
The active fraction that displayed the highest activity from HPLC-purified peptides was selected for LC–MS/MS analysis to confirm the accurate peptide mass and putative disulphide bond location. Liquid chromatography and mass spectrometry analysis were performed as outlined in Swart et al. [51], with a few modifications. A Thermo Scientific Ultimate 3000 RSLC equipped with a C18 trap column (PepMap™; 100 Å pore size, 3 µm particle size, 0 75 µm × 20 mm) and a C18 analytical column (Waters nanoEase M/Z Peptide CSH C18 Column; 130 Å pore size, 1.7 µm particle size, 150 µm × 150 mm) were used for the LC analysis. The solvent system employed was loading: 2% acetonitrile/water containing 0.1% formic acid; Solvent A: water containing 0.1% formic acid and Solvent B: acetonitrile containing 0.1% formic acid. The samples were loaded onto the trap column using the loading solvent at a flow rate of 2 µL/min from a temperature-controlled autosampler set at 7 °C. Loading was performed for 5 min before the sample was eluted onto the analytical column. The flow rate was set to 0.3 µL/min, and the gradient was generated as follows: 5% to 85% Solvent B from 5 to 45 min, 85% to 5% from 45 to 75 min using Chromeleon non-linear gradient 5. Chromatography was performed at 50 °C, and the outflow was delivered to the mass spectrometer through a stainless-steel nano-bore emitter.
Mass spectrometry was performed using an Orbitrap Fusion mass spectrometer (Thermo Scientific, Waltham, Massachusetts) equipped with a Nanospray Flex ionisation source. Data were collected in positive mode with spray voltage set to 1.9 kV and ion transfer capillary set to 275 °C. Spectra were internally calibrated using polysiloxane ions at m/z = 445.12. The MS1 scans were performed using the orbitrap detector set at 120 000 resolution over the scan range of m/z 375–1500 with automatic gain control (AGC) target at 40,000 and a maximum injection time of 50 ms. Data were acquired in profile mode. The MS2 acquisitions were performed using monoisotopic precursor selection for ions with charges + 2 to + 9 with error tolerance set to approximately 10 ppm. Precursor ions were excluded from fragmentation once for a period of 60 s. Precursor ions were selected for fragmentation in higher-energy collisional dissociation (HCD) mode using the quadrupole mass analyser with HCD energy set to 30%. Fragment ions were detected with the Orbitrap mass analyser set to 30,000 resolution. The AGC target was set to 50,000 and the maximum injection time to 100 ms. The data were acquired in centroid mode. The LC–MS/MS data were processed and analysed using the MZmine 2 software [52] and pLink® 2 [53].
Minimum Inhibitory Concentration (MIC)
The minimum inhibitory concentrations (MICs) were determined by following the standard broth microdilution method [54]. Briefly, microplates (Greiner CELLSTAR® 96-well plate; Sigma-Aldrich) were loaded with 100 µL of two-fold serial dilutions of the HPLC-purified PlaX_Opt and MunX_Opt in sterile Milli-Q water (starting at 1 mg/mL) and 100 µL of BHI broth. Overnight cultures of L. monocytogenes were diluted in BHI broth to obtain A625 of 0.08–0.1 (representing 1 × 108 CFU/mL), of which 10 µL was added to the respective wells. A positive growth control (L. monocytogenes in BHI broth without peptide treatment), a negative control (sterile BHI broth) and a sterility control (BHI broth containing peptide) were included for each MIC assay. The micro-plates were incubated at 37 °C for 8 h with agitation, whereafter the susceptibility of the organisms to the peptides was determined by measuring the A625 before (t = 0) and after incubation (t = 8) using an xMark™ Microplate Spectrophotometer (Bio-Rad, San Francisco, USA). The percentage inhibition was determined as the amount of bacteriocin that inhibited growth by at least 90%. The MIC values were reported as the means of two biological repeats in triplicate technical repeats.
Statistical Analysis
All the data were presented as mean ± standard deviation (SD). For statistical analysis, t-tests and one-way analysis of variance (ANOVA) tests were performed, followed by Tukey’s multiple comparison test. Differences with a p < 0.05 were considered statistically significant.
Nucleotide Sequence Accession Numbers
The codon-optimised gene sequences of plantaricin 423 and mundticin ST4SA were submitted to GenBank and assigned the accession numbers OQ703933 and OQ703932, respectively.
Results
Several industries will benefit from the cost-effective production of AMPs, with possible applications ranging from pharmaceuticals to food preservation. This study aimed to develop an improved expression system for producing class IIa bacteriocins in S. cerevisiae. The strategies to improve the production of two bacteriocins included codon optimisation, different secretion signals, and a strong constitutive promotor. Gene cassettes containing various combinations of secretion signals and bacteriocin gene variants were cloned into S. cerevisiae Y294, and the strains were evaluated for peptide production and activity.
Gene Variants and Strain Construction
In this study, the plaA, plaA_Opt, munST4SA and munST4SA_Opt genes were cloned into S. cerevisiae Y294 to express plantaricin 423 and mundticin ST4SA, respectively (Table 1). The codon optimisation was used to replace rare codons in the DNA sequence with codons preferred by S. cerevisiae, thereby increasing the codon frequency (Online Resource 5). The varying distribution of preferred codons in organisms (also known as the codon usage bias) directly affects the translation efficiency of recombinant genes. Based on the codon usage bias for a specific host, different indices can be used to predict the expression of individual gene sequences [34]. For example, the codon bias index (CBI) measures the directional codon bias, i.e. the extent to which a gene uses a subset of ‘optimal’ codons. Alternatively, the codon adaptation index (CAI) measures the usage of preferred codons in a reference set without categorising codons as optimal or non-optimal. Codon optimisation alters the codon usage pattern using synonymous codons, but this strategy does not work for all genes as some of the intrinsic aspects of this technique remain unclear.
The CBI (codon bias index) for plaA increased from 0.14 to 0.23 after codon optimisation and for munST4SA from 0.05 to 0.59 (Online Resource 6). While the CAI (codon adaption index) predicted a higher increase in expression for plaA_Opt than munST4SA_Opt, the CBI results predicted an 11.8-fold higher expression for munST4SA_Opt and only 1.6-fold for plaA_Opt. Plantaricin 423 peptides produced by S. cerevisiae strains Y294[MFα1-PlaX], Y294[MFα1-PlaX_Opt], Y294[XYN-PlaX] and Y294[XYN-PlaX_Opt] were refered to as MFα1-PlaX, MFα1-PlaX_Opt, XYN-PlaX and XYN-PlaX_Opt, respectively. Mundticin ST4SA peptides produced by S. cerevisiae strains Y294[MFα1-MunX], Y294[MFα1-MunX_Opt], Y294[XYN-MunX] and Y294[XYN-MunX_Opt] were referred to as MFα1-MunX, MFα1-MunX_Opt, XYN-MunX and XYN-MunX_Opt, respectively. The MFα1 and XYNSEC (XYN) secretion signals were indicated within the different gene variants. The Y294[BBH1] control contains an ENO1 cassette with no secretion signal, the Y294[BBH4] control contains the ENO1 cassette with the XYNSEC secretion, and Y294[MR] contains the ENO1 cassette with the MFα1 secretion signal.
Screening for Antimicrobial Activity
The recombinant S. cerevisiae strains were evaluated for the production of the plantaricin 423 and mundticin ST4SA peptides using L. monocytogenes EDG-e as the indicator organism. In the agar-overlay assay, clear and distinct inhibition zones were observed around the recombinant strains containing either the native and/or codon-optimised peptides with the MFα1 secretion signal (Fig. 1a). Little activity was observed for the strains containing the XYNSEC secretion signal, while no activity was observed for the negative control strains. The agar well-diffusion assays with the CFSs confirmed extracellular anti-listerial activity (clear inhibition zones) for strains containing the MFα1 secretion signal (Fig. 1b). No activity was observed in the supernatant of strains containing the XYNSEC secretion signal, suggesting that the peptides are not secreted into the supernatant.
Antimicrobial activity of the recombinant S. cerevisiae strains against L. monocytogenes EDG-e. a Agar-overlay assays with yeast strains and b agar well-diffusion assays with CFS from strains producing plantaricin 423 (PlaX) peptide (left) or mundticin ST4SA (MunX) peptide (right). The negative controls are BBH1, BBH4 and MR
The results obtained for the agar-overlay and agar well-diffusion assays are summarised in Online Resource 7. The agar-overlay assay showed larger inhibition zones for strains containing the MFα1 secretion signal than those with the XYNSEC secretion signal. The [MFα1-MunX_Opt] and [MFα1-MunX] strains displayed average inhibition zones of 41.3 ± 0.5 mm and 40.3 ± 0.5 mm, respectively, while the [MFα1-PlaX-Opt] and [MFα1-PlaX] strains had average inhibition zones of 27 ± 0.0 mm and 25.7 ± 0.9 mm. In contrast, the [XYN-MunX_Opt], [XYN-MunX], [XYN-PlaX_Opt] and [XYN-PlaX] strains displayed inhibition zones of 6 ± 0.0 mm to 8.3 ± 0.5 mm. The antimicrobial activity for PlaX thus increased by more than 70% and by more than 80% for MunX when expressed with the MFα1 secretion signal relative to the XYNSEC secretion signal. Whereas the CFS from strains containing MFα1 produced inhibition zones of 11.5 ± 2.1 mm for [MFα1-MunX] to 17.5 ± 3.2 mm for [MFα1-PlaX], no activity was observed in the supernatant of strains containing the XYNSEC secretion signal. After treatment with trypsin, the supernatants displayed reduced activity, thus confirming that the activity was linked to protein species and not secreted metabolites (data not shown).
Quantifying Bacteriocin Activity
Bacteriocin activity assays were performed on the supernatant of strains containing the MFα1 secretion signal to quantify their antimicrobial activity (in AU/mL) over 72 h of cultivation (Fig. 2). Antimicrobial activity was observed for all the AMP-producing strains; both the native and codon-optimised PlaX strains reached 320 ± 0.0 AU/mL after 72 h, whereas the native and codon-optimised MunX strains yielded 67 ± 18.86 AU/mL and 533 ± 150.85 AU/mL, respectively, representing a significant eightfold increase (p < 0.05). The DCWs of the AMP-producing and negative control strains were comparable, indicating that cell growth was not affected by bacteriocin production.
SDS-PAGE Analysis
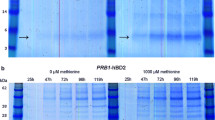
Tricine SDS-PAGE analysis of 20-fold concentrated supernatants indicated the presence of protein species that correspond to the size of plantaricin 423 (PlaX, 3.9 kDa) and mundticin ST4SA (MunX, 4.2 kDa) in the respective MFα1-containing strains (Fig. 3a, d). No similar size peptides were detected for the negative control or XYNSEC-containing strains. The tricine SDS-PAGE gels were subjected to anti-listerial agar overlay assays, which showed clear zones for the MFα1-containing strains (Fig. 3b, e). The inhibition zones corresponded to the respective peptide size of plantaricin 423 and mundticin ST4SA (Fig. 3c, f).
Tricine SDS-PAGE analysis of equal volumes of 20-fold concentrated supernatant containing recombinant a–c plantaricin 423 (PlaX) or d–f mundticin ST4SA (MunX) peptides. Images a and d represent silver-stained gels; b and e are overlay gels with L. monocytogenes; c and f represent the superimposed gels. Arrows indicate the recombinant PlaX and MunX peptides, respectively. The MFα1 and XYN controls refer to the supernatant of the Y294[MR] and Y294[BBH4] control strains containing the MFα1 and XYNSEC secretion signals, respectively
Based on the results obtained from the antimicrobial activity assays and tricine SDS-PAGE analysis, the yeast strains producing the peptides under the control of the MFα1 secretion signal were selected for further evaluation.
Stability Assay
The peptides containing the codon-optimised and native nucleotide sequences retained their stability after exposure to temperatures ranging from 4 to 100 °C and a pH of 2.0 to 10.0 (Online Resource 8). For all the strains, antimicrobial activity correlated with an increase in temperature. The ANOVA and Tukey tests revealed significant differences for PlaX_Opt treated at 100 °C relative to treatments at 4 °C, 30 °C and 37 °C, whereas MFα1-PlaX had significantly higher activity at temperatures above 60 °C. The antimicrobial activity of MunX_Opt and MFα1-MunX exposed to temperatures above 80 °C was significantly higher than at the lower temperatures. These results compared well with previous reports on the temperature and pH stability of the native peptides [14].
SEM Analysis
After the L. monocytogenes cells were exposed for 18 h to the concentrated CFS of the Y294[MFα1-PlaX_Opt] and [MFα1-MunX_Opt] strains, changes in the bacterial cell morphology were clearly visible with SEM (Fig. 4). The untreated L. monocytogenes cells remained intact and the cell walls appeared smooth (Fig. 4a). After treatment with supernatant from Y294[MFα1-PlaX_Opt] (Fig. 4b) or [MFα1-MunX_Opt] (Fig. 4c), the cells appeared shrivelled, pores formed in the cell wall and intracellular debris were released from the cells, signifying that the L. monocytogenes cell membranes were disrupted. After overnight incubation, no growth was observed for the samples treated with either the PlaX_Opt or MunX_Opt peptides, while the untreated sample displayed growth (data not shown).
SEM images of L. monocytogenes cells after 18 h a without treatment, b treated with PlaX_Opt and c treated with MunX_Opt. The untreated L. monocytogenes cells appear smooth and intact, but shrivelled after treatment with PlaX_Opt or MunX_Opt. The arrows indicate intracellular debris released from cells due to pore formation in the cell wall
Nano-LC–MS and MS/MS Analysis
The recombinant peptides were harvested from S. cerevisiae Y294[MFα1-PlaX_Opt] and Y294[MFα1-MunX_Opt], respectively, TCA-acetone precipitated and purified using HPLC (with a C8 analytical column). The collected fractions were spot-tested for anti-listerial activity (Figs. 5a and 6a). The HPLC-purified fractions 8–12 of plantaricin 423 and fractions 8–17 of mundticin ST4SA displayed antimicrobial activity. Fractions with the highest anti-listerial activity (fraction 9 of plantaricin 423 and fraction 8 of mundticin ST4SA were further analysed with nano-LC–MS/MS (Figs. 5b–d and 6b–e).
HPLC purification, accurate mass determination and peptide sequence confirmation of PlaX_Opt. a HPLC fractionation (C8 column) of PlaX_Opt with anti-listerial activity identified in fractions 8–12, with fraction 9 displaying the highest activity and subsequently analysed with LC–MS/MS. Monoisotopic ions (red arrows) were observed for PlaX_Opt carrying b + 5 charges ([M + 5H]+5 with an expected m/z 786.7493) and c + 4 charges ([M + 4H]+4 with an expected m/z 983.1848). d Collision-induced peptide fragmentation spectra confirmed the PlaX_Opt peptide sequence and the two disulphide bonds represented as sections of no fragmentation
HPLC purification, accurate mass determination and peptide sequence confirmation of MunX_Opt. a HPLC fractionation (C8 column) of MunX_Opt with anti-listerial activity identified in fractions 8–17, with fraction 8 displaying the greatest antimicrobial activity and subsequently analysed with LC–MS/MS. Monoisotopic ions (red arrows) were observed for MunX_Opt carrying b + 6 charges ([M + 6H]+6 with an expected m/z 715.1898), c + 5 charges ([M + 5H]+5 with an expected m/z 858.0264 and d + 4 charges ([M + 4H]+4 with an expected m/z 1072.2811). e Collision-induced peptide fragmentation spectra confirmed the peptide sequence of MunX_Opt and single disulphide bond represented as the section of no fragmentation
Nano-LC–MS performed on the HPLC-purified active fraction of PlaX_Opt confirmed the presence of a peptide with a mass corresponding to that of the mature plantaricin 423 with two disulphide bonds from the isotopic envelopes of the [M + 5H]+5 and [M + 4H]+4 species (Fig. 5b, c). The accurate mass measurements closely agree with the theoretical monoisotopic mass of plantaricin 423 (3928.710 Da) (Online Resource 9). A mass corresponding to the mature mundticin ST4SA was confirmed from the isotopic envelopes of the [M + 6H]+6, [M + 5H]+5 and [M + 4H]+4 species (Fig. 6b–d). The accurate mass measurements for MunX_Opt also corresponded to the theoretical monoisotopic mass of mundticin ST4SA (4285.095 Da), equivalent to forming one disulphide bond (Online Resource 9). The monoisotopic parent ion from the peptide envelopes was fragmented and analysed with tandem mass spectrometry (Figs. 5d and 6e). Based on the fractionation patterns, the peptide sequences could accurately be determined with the correct disulphide bond formation, thereby confirming the identities of the recombinantly produced plantaricin 423 and mundticin ST4SA (with a mass error of 10 ppm). The ions observed from the fractionation patterns of plantaricin 423 and mundticin ST4SA are summarised in Online Resource 10 and Online Resource 11, respectively.
MICs and Yields of HPLC-Purified Peptides
All the active fractions of HPLC-purified PlaX_Opt and MunX-Opt (on the C8 column) were collected and combined, freeze-dried and analytically weighed to determine the peptide yields, with 18.4 ± 3.40 and 20.9 ± 1.59 mg/L obtained for PlaX_Opt and MunX_Opt, respectively (Table 3). Vermeulen et al. [25] reported 12.4 mg/L of purified mundticin and theoretically 12 mg/L of plantaricin produced in E. coli. The yields in S. cerevisiae were 40.67% and 34.78% higher for active MunX_Opt and PlaX_Opt than E. coli.
The MICs of the native plantaricin and mundticin against L. monocytogenes strain EDG-e were reported to be 12 µM and 10 µM, respectively [55]. These MICs are lower than that observed for the recombinant PlaX_Opt and MunX_Opt against the same strain in this study. However, the native peptides were not 95% pure, and a direct comparison should be treated with caution. The purity of the HPLC-purified peptides in this study was assumed to be > 95% as determined by tricine SDS-PAGE analysis and densitometry (Online Resource 12 and Online Resource 13). Moreover, the respective Y294[MFα1-PlaX_Opt] and Y294[MFα1-MunX_Opt] strains produced 3.96-fold more PlaX_Opt and 44.95-fold more MunX_Opt than required for MIC against L. monocytogenes.
Discussion
In this study, a yeast expression system for the production of bacteriocins in S. cerevisiae was developed and optimised using plantaricin 423 and mundticin ST4SA as benchmark peptides. Codon-optimised and native variants of the plaA and munST4SA genes, encoding the mature plantaricin 423 and mundticin ST4SA peptides, respectively, were cloned in-frame into yeast expression plasmids containing either the MFα1 or XYNSEC secretion signals under the transcriptional control of the ENO1 promotor and terminator. The plasmids were introduced into S. cerevisiae Y294, and the various recombinant strains were evaluated for peptide production and antimicrobial activity.
Results from the agar overlay assays indicated that the inhibition zones observed around the recombinant S. cerevisiae strains containing the MFα1 secretion signal were more than 80% larger than for strains containing the XYNSEC secretion signal. However, the agar well-diffusion assays did not indicate activity in the supernatant from the XYNSEC strains, suggesting that the peptides were not secreted extracellularly. This was confirmed when no extracellular proteins were detected for the XYNSEC strains by tricine SDS-PAGE. Although the XYNSEC secretion signal has been used successfully for the secretion of large proteins in S. cerevisiae (such as amylases) [41], our results showed that the MFα1 secretion signal was superior for the secretion of the much smaller plantaricin 423 and mundticin ST4SA peptides. It is plausible that the MFα1 secretion signal was evolutionarily adapted to facilitate improved secretion of the small MFα mating type peptide [37].
Codon optimisation can enhance gene expression as it alters the codon bias of a foreign protein (bacterial antimicrobial peptides in this case) to match the codon bias of the host organism (e.g. S. cerevisiae), but positive results are not guaranteed [56]. The bacteriocin activity assays indicated an eightfold increase in activity for the peptide encoded by the codon-optimised mundticin gene compared to the native gene, which could result from enhanced gene expression. However, the plantaricin 423 peptides produced by the native and codon-optimised genes showed no significant difference in activity. From Online Resource 5, it is clear that the overall relative frequency of codons increased after codon optimisation for both genes, but the codon bias index (CBI) increase for munST4SA_Opt was higher than for plaA_Opt relative to the native genes. The lower increase in the CBI for plaA could explain why no significant difference in the activity of the plantaricin peptides produced by the native and codon-optimised genes was observed. In contrast, codon optimisation of munST4SA substantially increased expression in S. cerevisiae.
The activity and mode of action of the recombinant peptides were confirmed with SEM analysis, which clearly showed shrivelled bacterial cells and leaking cytoplasm when exposed to the AMPs. The stability of the recombinant peptides was retained after exposure to different pH ranges and temperatures (Online Resource 8), but an increase in activity was observed for both recombinant peptides at higher temperatures. This could result from the degradation of endogenous-produced proteases and proteolytic enzymes at high temperatures, which do not affect the bacteriocins.
The mass spectrometry results confirmed the identities of the recombinant peptides with the correct conformation of disulphide bonds. Furthermore, significant yields were obtained for both peptides (18.4 mg/L plantaricin 423 and 20.9 mg/L mundticin ST4SA). One of the major limiting factors of bacteriocin production is low yields of purified peptides. In a study by Jiang et al. [57], yields of 2–2.5 mg/L plantaricin NC8α and 1.5–2 mg/L plantaricin NC8β were obtained from heterologous expression in E. coli. Meng et al. [58] expressed plantaricin Pln1 as a fusion protein with thioredoxin in E. coli and obtained 100–110 mg/L fused protein and 9–11 mg/L cleaved peptide. Furthermore, the heterologous co-production of enterocin A and pediocin PA-1 in Lactococcus lactis resulted in 27 ng/L and 406 ng/L peptide, respectively [59].
The increased yields and simple purification strategy employed in this study support the notion that yeast may be a better candidate for the expression of plantaricin 423 and mundticin ST4SA. There was no need for fusion proteins as the peptides are not toxic towards the yeast, and the bacteriocins were efficiently secreted into the supernatant, simplifying the harvesting of the peptides. Production of the peptides in E. coli requires lysis of the bacterial cells to harvest the peptides, which does not guarantee a 100% peptide recovery rate [25]. Furthermore, the yeast expression system does not require chemical induction (such as IPTG) for the expression and production of bacteriocins, nor does it require protease-mediated liberation of the bacteriocin from a fusion partner [25], which can become costly with the large-scale production of the peptides. Even though bacterial systems can be developed for constitutive peptide production and secretion, antimicrobial peptides can become toxic to bacterial cells. The lack of toxicity against yeast cells, coupled with different options for secretion and constitutive expression, renders yeast advantageous for antimicrobial peptide production. After IMAC purification, mature peptide cleavage, HPLC-purification and lyophilisation, only 12.4 mg/L active mundticin ST4SA was recovered from E. coli by Vermeulen et al. [25], compared to 20.9 mg/L mundticin ST4SA produced by S. cerevisiae. The yields and purification strategy in this study thus offer significant advantages compared to production in E. coli.
The S. cerevisiae expression system constructed in this study also outperformed the expression systems reported by Schoeman et al. [30] and Van Reenen et al. [31]. This could be due to codon optimisation of the bacteriocin genes for expression in S. cerevisiae, and the use of the constitutive ENO1 promotor and terminator (instead of the inducible ADH1 promotor and terminator). However, the production of the peptides in fed-batch bioreactors under controlled conditions should be investigated as this could yield improved biomass production and potentially higher peptide yields. For example, fed-batch fermentation with a 7 g/L/h stable sucrose feeding rate significantly enhanced the production of pediocin SM-1 by Pediococcus pentosaceus compared to batch culture [60].
Pichia pastoris has always been regarded as the frontrunner for producing bacteriocins from yeast, as the yeast can achieve high concentrations of biomass that result in high protein titers. However, yields reported for purified bacteriocins from P. pastoris are not much higher than those obtained from S. cerevisiae in this study. Li et al. [61] expressed a hybrid bacteriocin (EF-1) derived from plantaricin E and plantaricin F in P. pastoris and obtained 32.65 mg/L of HPLC-purified peptide, while Gutiérrez et al. [62] achieved 22.8 mg/L pure enterocin P from P. pastoris. The recombinant expression of bacteriocins in P. pastoris mainly involved the AOX promotor, which requires induction by methanol, which involves additional costs and complicates purification and downstream applications.
When comparing the activity of the recombinant plantaricin 423 and mundticin ST4SA-producing yeast with the native bacteriocin-producing strains, the MIC of the recombinant S. cerevisiae strains were tenfold lower for plantaricin 423 and 92-fold lower for mundticin [55]. Rich and complex growth media is required for the native hosts that renders production expensive and peptide purification difficult. As a result, the purity, and consequently the specific activity, of the native peptides was lower than that achieved for the recombinant peptides produced in this study. The advantages of our expression system include the more consistent production of peptides and simplified purification. Bacteriocin expression by the native host is highly regulated, which can result in inconsistent production [19, 20]. It has been hypothesised that quorum sensing is the primary mode of class IIa bacteriocin regulation, as gene expression depends on producer cell density [63]. Other factors that could influence and further complicate bacteriocin production include temperature and divalent metal cation concentrations [19, 20, 64]. In contrast, the yeast expression system produces bacteriocins constitutively in an active form on relatively cheap media. Furthermore, a simplified purification process was used when compared to the production of plantaricin and mundticin from the native hosts [55]. Due to the simplified S. cerevisiae expression system developed in this study, bio-mining and characterisation of novel peptides, as well as peptides that are highly regulated by their native hosts, could be more achievable.
Although improved expression of bacteriocins by the S. cerevisiae laboratory strain was demonstrated as a proof of concept, expression of the peptides in an industrial S. cerevisiae strain should be investigated for commercial application and up-scaled peptide production. Industrial S. cerevisiae strains are better suited to reach high cell densities and withstand harsh industrial conditions such as high temperature, pH and ethanol concentrations. Furthermore, an optimised purification process needs to be developed for the large-scale production of the peptides, as this remains a limiting step.
Conclusions
The plantaricin 423 and mundticin ST4SA bacteriocins were successfully expressed in S. cerevisiae, and the recombinant peptides retained their activity and stability. Different secretion signals and the impact of codon optimisation for expressing the bacteriocin genes in S. cerevisiae were investigated. Higher anti-listerial activity was observed for the yeast strain containing the codon-optimised munST4SA gene, while no significant difference in activity was observed for the strains containing the codon-optimised plaA gene compared to the native bacterial genes. The fusion of the bacteriocin genes to the MFα1 secretion signal allowed for the effective secretion of the bacteriocins into the yeast supernatant, while the peptides apparently remained bound to the yeast cell wall with the XYNSEC secretion signal. Following HPLC-purification, the recombinant S. cerevisiae strains yielded 20.9 mg/L of mundticin ST4SA and 18.4 mg/L of plantaricin 423. This is a marked improvement over the yields previously reported for plantaricin 423 and mundticin ST4SA in E. coli expression systems, and they required a much simpler purification process.
This study highlights the potential of using S. cerevisiae as a host for the heterologous production of antimicrobial peptides. The expression system described in this study could simplify the expression and characterisation of novel peptides, including antimicrobial peptides from other prokaryotes and eukaryotes from underrepresented classes. In addition, future studies should investigate the up-scaled production of the peptides by S. cerevisiae in bioreactors for improved yields.
Data Availability
The data sets generated in this study are available on request from the corresponding author.
References
Hwang PM, Vogel HJ (1998) Structure-function relationships of antimicrobial peptides. Biochem Cell Biol 76(2–3):235–246. https://doi.org/10.1139/bcb-76-2-3-235
Lohans CT, Vederas JC (2012) Development of class IIa bacteriocins as therapeutic agents. Int J Microbiol 1–13. https://doi.org/10.1155/2012/386410
Kumariya R, Garsa AK, Rajput YS, Sood SK, Akhtar N, Patel S (2019) Bacteriocins: classification, synthesis, mechanism of action and resistance development in food spoilage causing bacteria. Microb Pathog 128:171–177. https://doi.org/10.1016/j.micpath.2019.01.002
Yi Y, Li P, Zhao F, Zhang T, Shan Y, Wang X, Liu B, Chen Y, Zhao X, Lü X (2022) Current status and potentiality of class II bacteriocins from lactic acid bacteria: structure, mode of action and applications in the Food Industry. Trends Food Sci Technol 120:387–401. https://doi.org/10.1016/j.tifs.2022.01.018
Alvarez-Sieiro P, Montalbán-López M, Mu D, Kuipers OP (2016) Bacteriocins of lactic acid bacteria: extending the family. Appl Microbiol Biotechnol 100:2939–2951. https://doi.org/10.1007/s00253-016-7343-9
Wu X, Ju X, Du L, Wang L, He R, Chen Z (2020) The Man-PTS subunit IIC is responsible for the sensitivity of Listeria monocytogenes to durancin GL. Food Sci Nutr 8(1):150–161. https://doi.org/10.1002/fsn3.1285
Kjos M, Salehian Z, Nes IF, Diep DB (2010) An extracellular loop of the mannose phosphotransferase system component IIC is responsible for specific targeting by class IIA bacteriocins. J Bacteriol 192(22):5906–5913. https://doi.org/10.1128/jb.00777-10
Ng ZJ, Zarin MA, Lee CK, Tan JS (2020) Application of bacteriocins in food preservation and infectious disease treatment for humans and livestock: a review. RSC Adv 10(64):38937–38964. https://doi.org/10.1039/d0ra06161a
Gumienna M, Górna B (2021) Antimicrobial food packaging with biodegradable polymers and bacteriocins. Molecules 26(12):3735. https://doi.org/10.3390/molecules26123735
Yap P, MatRahim N, AbuBakar S, Lee HY (2021) Antilisterial potential of lactic acid bacteria in eliminating Listeria monocytogenes in host and ready-to-eat food application. Microbiol Res 12(1):234–257. https://doi.org/10.3390/microbiolres12010017
Darbandi A, Asadi A, Ari MM, Ohadi E, Talebi M, Zadeh MH, Emamie AD, Ghanavati R, Kakanj M (2021) Bacteriocins: properties and potential use as antimicrobials. J Clin Lab Anal 36(1):e24093. https://doi.org/10.1002/jcla.24093
Cintas LM, Casaus P, Fernández MF, Hernández PE (1998) Comparative antimicrobial activity of enterocin L50, pediocin PA-1, Nisin A and lactocin S against spoilage and foodborne pathogenic bacteria. Food Microbiol 15(3):289–298. https://doi.org/10.1006/fmic.1997.0160
Millette M, Cornut G, Dupont C, Shareck F, Archambault D, Lacroix M (2008) Capacity of human nisin- and pediocin-producing lactic acid bacteria to reduce intestinal colonization by vancomycin-resistant Enterococci. Appl Environ Microbiol 74(7):1997–2003. https://doi.org/10.1128/aem.02150-07
Van Reenen CAA, Dicks LMT, Chikindas ML (1998) Isolation, purification and partial characterization of plantaricin 423, a bacteriocin produced by Lactobacillus plantarum. J Appl Microbiol 84:1131–1137. https://doi.org/10.1046/j.1365-2672.1998.00451.x
Granger M, van Reenen CAA, Dicks LMT (2008) Effect of gastrointestinal conditions on the growth of Enterococcus mundtii ST4SA, and production of bacteriocin ST4SA recorded by real-time PCR. Int J Food Microbiol 123:277–280. https://doi.org/10.1016/j.ijfoodmicro.2007.12.009
Van Zyl WF, Deane SM, Dicks LMT (2019) Bacteriocin production and adhesion properties as mechanisms for the anti-listerial activity of Lactobacillus plantarum 423 and Enterococcus mundtii ST4SA. Benef Microbes 10(3):329–349. https://doi.org/10.3920/bm2018.0141
Mesa-Pereira B, Rea MC, Cotter PD, Hill C, Ross RP (2018) Heterologous expression of biopreservative bacteriocins with a view to low cost production. Front Microbiol 9. https://doi.org/10.3389/fmicb.2018.01654
Nes IF, Diep DB, Håvarstein LS, Brurberg MB, Eijsink V, Holo H (1996) Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Leeuwenhoek 70(2–4):113–128. https://doi.org/10.1007/bf00395929
Diep DB, Axelsson L, Grefsli C, Nes IF (2000) The synthesis of the bacteriocin sakacin A is a temperature-sensitive process regulated by a pheromone peptide through a three-component regulatory system. Microbiology (Reading) 146(Pt 9):2155–2160. https://doi.org/10.1099/00221287-146-9-2155
Gursky LJ, Martin NI, Derksen DJ, van Belkum MJ, Kaur K, Vederas JC, Stiles ME, McMullen LM (2006) Production of piscicolin 126 by Carnobacterium maltaromaticum UAL26 is controlled by temperature and induction peptide concentration. Arch Microbiol 186:317–325. https://doi.org/10.1007/s00203-006-0147-z
Naghmouchi K, Fliss I, Drider D, Lacroix C (2008) Pediocin PA-1 production during repeated-cycle batch culture of immobilized Pediococcus acidilactici UL5 cells. J Biosci Bioeng 105(5):513–517. https://doi.org/10.1263/jbb.105.513
Cao J, de la Fuente-Nunez C, Ou R, Torres M, Pande S, Sinskey A, Lu T (2018) Yeast-based synthetic biology platform for antimicrobial peptide production. ACS Synth Biol 7(3):896–902. https://doi.org/10.1021/acssynbio.7b00396
Rosano GL, Ceccarelli EA (2014) Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol 5:172. https://doi.org/10.3389/fmicb.2014.00172
Li Y (2011) Recombinant production of antimicrobial peptides in Escherichia coli: a review. Protein Expr Purif 80(2):260–267. https://doi.org/10.1016/j.pep.2011.08.001
Vermeulen RR, Van Staden AD, Dicks LMT (2020) Heterologous expression of the class IIa bacteriocins, plantaricin 423 and mundticin ST4SA, in Escherichia coli using green fluorescent protein as a fusion partner. Front Microbiol 11:1634. https://doi.org/10.3389/fmicb.2020.01634
Sevier C, Kaiser C (2002) Formation and transfer of disulphide bonds in living cells. Nat Rev Mol Cell Biol 3:836–847. https://doi.org/10.1038/nrm954
Ahmad M, Hirz M, Pichler H, Schwab H (2014) Protein expression in Pichia pastoris: recent achievements and perspectives for heterologous protein production. Appl Microbiol Biotechnol 98(12):5301–5317. https://doi.org/10.1007/s00253-014-5732-5
Nielsen J (2013) Production of biopharmaceutical proteins by yeast. Bioengineered 4(4):207–211. https://doi.org/10.4161/bioe.22856
Kim H, Yoo SJ, Kang HA (2015) Yeast synthetic biology for the production of recombinant therapeutic proteins. FEMS Yeast Res 15(1):1–16. https://doi.org/10.1111/1567-1364.12195
Schoeman H, Vivier M, du Toit M, Dicks LMT, Pretorius I (1999) The development of bactericidal yeast strains by expressing the Pediococcus acidilactici pediocin gene (pedA) in Saccharomyces cerevisiae. Yeast 15(8):647–656. https://doi.org/10.1002/(SICI)1097-0061(19990615)15:8%3c647::AID-YEA409%3e3.0.CO;2-5
Van Reenen CA, Chikindas ML, Van Zyl WH, Dicks LMT (2003) Characterization and heterologous expression of a class IIa bacteriocin, plantaricin 423 from Lactobacillus plantarum 423. Saccharomyces cerevisiae Int J Food Microbiol 81(1):29–40. https://doi.org/10.1016/S0168-1605(02)00164-2
Basanta A, Herranz C, Gutiérrez J, Criado R, Hernández PE, Cintas LM (2009) Development of bacteriocinogenic strains of Saccharomyces cerevisiae heterologously expressing and secreting the leaderless enterocin L50 peptides L50A and L50B from Enterococcus faecium L50. Appl Environ Microbiol 75(8):2382–2392. https://doi.org/10.1128/aem.01476-08
Nguyen TPA, Nguyen TTM, Nguyen NH, Nguyen TN, Dang TTP (2020) Application of yeast surface display system in expression of recombinant pediocin PA-1 in Saccharomyces cerevisiae. Folia Microbiol 65:955–961. https://doi.org/10.1007/s12223-020-00804-6
Fox J, Erill I (2010) Relative codon adaptation: a generic codon bias index for prediction of gene expression. DNA Res 17(3):185–196
Kleiner-Grote GRM, Risse JM, Friehs K (2018) Secretion of recombinant proteins from E. coli. Eng Life Sci 18(8):532–550. https://doi.org/10.1002/elsc.201700200
Myburgh MW, Rose SH, Viljoen-Bloom M (2020) Evaluating and engineering Saccharomyces cerevisiae promoters for increased amylase expression and bioethanol production from raw starch. FEMS Yeast Res 20(6). https://doi.org/10.1093/femsyr/foaa047
Kurjan J, Herskowitz I (1982) Structure of a yeast pheromone gene (MFα): a putative α-factor precursor contains four tandem copies of mature α-factor. Cell 30:933–943. https://doi.org/10.1016/0092-8674(82)90298-7
Van Rooyen R, Hahn-Hägerdal B, La Grange D, van Zyl WH (2005) Construction of cellobiose-growing and fermenting Saccharomyces cerevisiae strains. J Biotechnol 120(3):284–295. https://doi.org/10.1016/j.jbiotec.2005.06.013
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Njokweni A, Rose SH, van Zyl WH (2012) Fungal β-glucosidase expression in Saccharomyces cerevisiae. J Mol Microbiol Biotechnol 39:1445–1452. https://doi.org/10.1007/s10295-012-1150-9
Cripwell RA, Rose S, Viljoen-Bloom M, van Zyl WH (2019) Improved raw starch amylase production by Saccharomyces cerevisiae using codon optimisation strategies. FEMS Yeast Res 19(2). https://doi.org/10.1093/femsyr/foy127
Cho KM, Yoo YJ, Kang HS (1999) δ-Integration of endo/exoglucanase and β-glucosidase genes into the yeast chromosomes for direct conversion of cellulose to ethanol. Enzyme Microb Technol 25:23–30. https://doi.org/10.1016/S0141-0229(99)00011-3
Holder I, Boyce S (1994) Agar well diffusion assay testing of bacterial susceptibility to various antimicrobials in concentrations non-toxic for human cells in culture. Burns 20(5):426–429. https://doi.org/10.1016/0305-4179(94)90035-3
Baindara P, Chaudhry V, Mittal G, Liao LM, Matos CO, Khatri N, Franco OL, Patil PB, Korpole S (2016) Characterization of the antimicrobial peptide Penisin, a class Ia novel lantibiotic from Paenibacillus sp. strain A3. Antimicrob Agents Chemother 60(1):580–591. https://doi.org/10.1128/aac.01813-15
Belguesmia Y, Bendjeddou K, Kempf I, Boukherroub R, Drider D (2020) Heterologous biosynthesis of five new class II bacteriocins from Lactobacillus paracasei CNCM I-5369 with antagonistic activity against pathogenic Escherichia coli strains. Front Microbiol 11:1198. https://doi.org/10.3389/fmicb.2020.01198
Daba H, Pandian S, Gosselin J, Simard R, Huang J, Lacroix C (1991) Detection and activity of a bacteriocin produced by Leuconostoc mesenteroides. Appl Environ Microbiol 57(12):3450–3455. https://doi.org/10.1128/aem.57.12.3450-3455.1991.50
Schägger H (2006) Tricine–SDS-PAGE. Nat Protoc 1(1):16–22. https://doi.org/10.1038/nprot.2006.4
O’Connell K, Stults JT (1997) Identification of mouse liver proteins on two-dimensional electrophoresis gels by matrix-assisted laser desorption/ionization mass spectrometry of in situ enzymatic digests. Electrophoresis 18:349–359. https://doi.org/10.1002/elps.1150180309
Meng D, Li W, Shi L, Lv Y, Sun X, Hu J, Fan Z (2019) Expression, purification and characterization of a recombinant antimicrobial peptide Hispidalin in Pichia pastoris. Protein Expr Purif 160:19–27. https://doi.org/10.1016/j.pep.2019.03.007
Reichhardt C, Ferreira JAG, Joubert L, Clemons KV, Stevens DA, Cegelski L (2015) Analysis of the Aspergillus fumigatus biofilm extracellular matrix by solid-state nuclear magnetic resonance spectroscopy. Eukaryot Cell 14(11):1064–1072. https://doi.org/10.1128/ec.00050-15
Swart PC, Russell VA, Vlok NM, Dimatelis JJ (2018) Early-ethanol exposure induced region-specific changes in metabolic proteins in the rat brain: a proteomics study. J Mol Neurosci 65:277–288. https://doi.org/10.1007/s12031-018-1097-z
Pluskal T, Castillo S, Villar-Briones A, Oresic M (2010) MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform 11:395. https://doi.org/10.1186/1471-2105-11-395
Chen ZL, Meng JM, Cao Y, Yin JL, Fang RQ, Fan SB, Liu C, Zeng WF, Ding YH, Tan D, Wu L, Zhou WJ, Chi H, Sun RX, Dong MQ, He SM (2019) A high-speed search engine pLink 2 with systematic evaluation for proteome-scale identification of cross-linked peptides. Nat Commun 10:3404. https://doi.org/10.1038/s41467-019-11337-z
Clinical and Laboratory Standards Institute (2012) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, approved standard. https://www.techstreet.com/mss/products/preview/1823012
Dreyer L, Smith C, Deane SM, Dicks LMT, Van Staden AD (2019) Migration of bacteriocins across gastrointestinal epithelial and vascular endothelial cells, as determined using in vitro simulations. Sci Rep 9(1):11481. https://doi.org/10.1038/s41598-019-47843-9
Gustafsson C, Govindarajan S, Minshull J (2004) Codon bias and heterologous protein expression. Trends Biotechnol 22(7):346–353. https://doi.org/10.1016/j.tibtech.2004.04.006
Jiang H, Li P, Gu Q (2016) Heterologous expression and purification of plantaricin NC8, a two-peptide bacteriocin against Salmonella spp. from Lactobacillus plantarum ZJ316. Protein Expr Purif 127:28–34. https://doi.org/10.1016/j.pep.2016.06.013
Meng F, Zhao H, Zhang C, Lu F, Bie X, Lu Z (2016) Expression of a novel bacteriocin - the plantaricin pln1 - in Escherichia coli and its functional analysis. Protein Expr Purif 119:85–93. https://doi.org/10.1016/j.pep.2015.11.008
Martínez JM, Kok J, Sanders JW, Hernández PE (2000) Heterologous coproduction of enterocin A and pediocin PA-1 by Lactococcus lactis: detection by specific peptide-directed antibodies. Appl Environ Microbiol 66(8):3543–3549. https://doi.org/10.1128/AEM.66.8.3543-3549.2000
Papagianni M, Papamichael EM (2014) Production of pediocin SM-1 by Pediococcus pentosaceus Mees 1934 in fed-batch fermentation: effects of sucrose concentration in a complex medium and process modelling. Process Biochem 49(12):2044–2048. https://doi.org/10.1016/j.procbio.2014.09.023
Li Z, Cheng Q, Guo H, Zhang R, Si D (2020) Expression of hybrid peptide EF-1 in Pichia pastoris, its purification, and antimicrobial characterization. Molecules 25(23):5538. https://doi.org/10.3390/molecules25235538
Gutiérrez J, Criado R, Martín M, Herranz C, Cintas LM, Hernández PE (2005) Production of enterocin P, an antilisterial pediocin-like bacteriocin from Enterococcus faecium P13. Pichia pastoris Antimicrob Agents Chemother 49(7):3004–3008. https://doi.org/10.1128/AAC.49.7.3004-3008.2005
Kuipers OP, de Ruyter PGGA, Kleerebezem M, de Vos WM (1998) Quorum sensing-controlled gene expression in lactic acid bacteria. J Biotechnol 64(1):15–21. https://doi.org/10.1016/s0168-1656(98)00100-x
Vermeulen RR, Deane S, Dicks LMT, Rohwer J, van Staden AD (2021) Manganese privation-induced transcriptional upregulation of the class IIa bacteriocin plantaricin 423 in Lactobacillus plantarum strain 423. Appl Environ Microbiol 87(21):e0097621. https://doi.org/10.1128/AEM.00976-21
Acknowledgements
Dr. Mare Vlok and Dr. Alicia Botes (CAF, Stellenbosch University) assisted with the LC-MS and SEM analysis, respectively. Prof. Marina Rautenbach (Biochemistry Department, Stellenbosch University) provided access to and assistance with the Micro Balance.
Funding
Open access funding provided by Stellenbosch University. This work was supported by the South African National Research Foundation (NRF) via Grants 86423 (WHvZ) and 118528 (MVB). Opinions communicated and all conclusions arrived at are those of the authors and are not necessarily to be attributed to the NRF.
Author information
Authors and Affiliations
Contributions
M.R., R.A.C., W.H.vZ and M.V.B. contributed to the study conception and design. Experimental design and execution were done by M.R.; R.R.V. and A.A.vS. assisted with data analysis. M.R. wrote the main manuscript text, and all the authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rossouw, M., Cripwell, R.A., Vermeulen, R.R. et al. Heterologous Expression of Plantaricin 423 and Mundticin ST4SA in Saccharomyces cerevisiae. Probiotics & Antimicro. Prot. 16, 845–861 (2024). https://doi.org/10.1007/s12602-023-10082-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-023-10082-6