Abstract
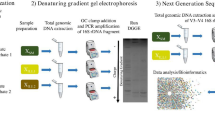
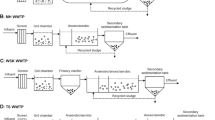
Microbial community DNA was extracted from activated sludge samples taken from a chemical bioflocculation process and a chemical coagulation process in Shanghai, China. 16S rDNA of ammonia-oxidizing bacteria (AOB)was amplified by nested polymerase chain reaction and fingerprinted by denaturing gradient gel electrophoresis for microbial structure analysis. The Shannon diversity index of each sample was determined. The results indicated that the microbial structure of AOB in chemical bioflocculation process was comparable at two operational conditions. The ammonia-oxidizing bacterial communities were similar in three channels of the chemical bioflocculation process and in three serial tanks in the chemical coagulation process at the same condition. The diversity of microbial structures in the chemical bioflocculation process was higher than in the chemical coagulation process, in which the microbial structure was similar to that in the influent. Although the microbial study provides insights to the nitrification removal, higher microbial diversity of AOB does not necessarily mean higher ammonia oxidization. Molecular analysis should be combined with chemical assays to optimize operational conditions.




Similar content being viewed by others
References
Cox GW (1979) General ecology experimental manual. Science, Beijing, pp 120–124
Limpiyakorn T, Shinohara Y, Kurisu F, Yagi O (2004) Distribution of ammonia-oxidizing bacteria in sewage activated sludge: analysis based on 16S rDNA sequence. Water Sci Technol 50:9–14
Muriel B, Wafa A, Vincent U, Thierry H (1999) DNA extraction from activated sludges. Curr Microbiol 38:315–319
Park H, Regan J, Noguera D (2002) Molecular analysis of ammonia-oxidizing bacterial populations in aerated–anoxic orbal processes. Water Sci Technol 46:273–280
Purohit H, Kapley A, Moharikar A, Narde G (2003) A novel approach for extraction of PCR-compatible DNA from activated sludge samples collected from different biological effluent treatment plants. J Microbiol Methods 52:315–323
Sam Brook J (1996) Molecular clone experimental manual. Science, Beijing, pp 919–929
Sofia A, Liu W, Ong S, Ng W (2004) In-situ characterization of microbial community in an A/O submerged membrane bioreactor with nitrogen removal. Water Sci Technol 50:41–48
Tsai Y, Olson B (1991) Rapid method for direct extraction of DNA from soil and sediments. Appl Environ Microbiol 57:1070–1074
Wanner J (1997) Microbial population dynamics in biological wastewater treatment plants. In: Cloete TE, Muyima NYO (eds) Microbial community analysis: the key to the design of biological wastewater treatment systems. IAQW, London, pp 35–59
Wu Z, Xia S, Yang D, Gao T (2003) Comparative research of chemical and biological flocculation process to municipal wastewater. Chongqing Environmental Sciences 25:12–14
Xia S, Wang F, Fu Y, Yang D, Ma X (2005) Biodiversity analysis of microbial community in the chem-bioflocculation treatment process. Biotechnol Bioeng 89(6):656–659
You S, Chuang S, Ouyang C (2003) Nitrification efficiency and nitrifying bacteria abundance in combined AS-RBC and A2O systems. Water Res 37:2281–2290
Zhang Z, Wei Q, Zhang G (2001) Technology and development trend of the enhanced primary treatment of municipal wastewater. Chongqing Environmental Sciences 23:46–49
Øvreas L, Forney L, Daae F, Torsvik V (1997) Distribution of bacterioplankton in meromictic lake Saelevannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol 63:3367–3373
Acknowledgements
This article is supported by the National High Technology Research and Development Program of China (863 Program) No. 2002AA601320 and the Shuguang Program of Shanghai.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xia, S., Shi, Y., Fu, Y. et al. DGGE analysis of 16S rDNA of ammonia-oxidizing bacteria in chemical–biological flocculation and chemical coagulation systems. Appl Microbiol Biotechnol 69, 99–105 (2005). https://doi.org/10.1007/s00253-005-0035-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-005-0035-5