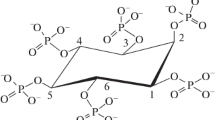
Abstract
Phytases are a special class of phosphatase that catalyze the sequential hydrolysis of phytate to less-phosphorylated myo-inositol derivatives and inorganic phosphate. Phytases are added to animal feedstuff to reduce phosphate pollution in the environment, since monogastric animals such as pigs, poultry, and fish are unable to metabolize phytate. Based on biochemical properties and amino acid sequence alignment, phytases can be categorized into two major classes, the histidine acid phytases and the alkaline phytases. The histidine acid phosphatase class shows broad substrate specificity and hydrolyzes metal-free phytate at the acidic pH range and produces myo-inositol monophosphate as the final product. In contrast, the alkaline phytase class exhibits strict substrate specificity for the calcium–phytate complex and produces myo-inositol trisphosphate as the final product. This review describes recent findings that present novel viewpoints concerning the molecular basis of phytase classification.




Similar content being viewed by others
References
Asada K, Tanaka K, Kasai Z (1969) Formation of phytic acid in cereal grains. Ann NY Acad Sci 165:801–814
Bajwa W, Meyhack B, Rudolph H, Schweingruber AM, Hinnen A (1984) Structural analysis of the two tandemly repeated acid phosphatase genes in yeast. Nucleic Acids Res 12:7721–7739
Barrientos L, Scott JJ, Murthy PP (1994) Specificity of hydrolysis of phytic acid by alkaline phytase from lily pollen. Plant Physiol 106:1489–1495
Baykov AA, Fabrichniy IP, Pohjanjoki P, Zyryanov AB, Lahti R (2000) Fluoride effects along the reaction pathway of pyrophosphatase: evidence for a second enzyme pyrophosphate intermediate. Biochemistry 39:11939–11947
Chang CW (1967) Study of phytase and fluoride effects in germinating corn seed. Cereal Chem 44:129–142
Cheryan M (1980) Phytic acid interactions in food systems. Crit Rev Food Sci Nutr 13:297–335
Choi YM, Suh HJ, Kim JM (2001) Purification and properties of extracellular phytase from Bacillus sp. KHU-10. J Protein Chem 20:287–292
Cosgrove DJ (1966) The chemistry and biochemistry of inositol polyphosphates. Rev Pure Appl Chem 16:209–215
Cosgrove DJ (1970) Inositol phosphate phosphatases of microbiological origin. Inositol phosphate intermediates in the dephosphorylation of the hexaphosphates of myo-inositol, scyllo-inositol, and d-chiro-inositol by a bacterial (Pseudomonas sp.) phytase. Aust J Biol Sci 23:1207–1220
Cosgrove DJ (1980) Phytases and intermediates in the dephosphorylation of P6-inositols by phytase enzymes. In: Cosgrove DJ (ed)Inositol phosphates: their chemistry, biochemistry, and physiology. (Studies in organic chemistry 4) Elsevere, Amsterdam, pp 85–105
Dossa E, Boquet PL (1985) Identification of the gene appA for the acid phosphatase (pH optimum 2.5) of Escherichia coli. Mol Gen Genet 200:68–73
Dossa J, Marck C, Boquet PL (1990) The complete nucleotide sequence of the Escherichia coli gene appA reveals significant homology between pH 2.5 acid phosphtase and glucose-1-phosphatase. J Bacteriol 172:5497–5500
Elliott S, Chang CW, Schweingruber ME, Schaller J, Rickli EE, Carbon J (1986) Isolation and characterization of the structural gene for secreted acid phosphatase from Schizosaccharomyces pombe. J Biol Chem 261:2936–2941
Englen AJ, Heeft FC van der, Randsdrop PH, Smit EL (1994) Simple and rapid determination of phytase activity. J AOAC Int 77:760–764
Eskin NAM, Wiebe S (1983) Changes in phytase activity and phytate during germination of two fababean cultivars. J Food Sci 48:270–271
Ethrlich KC, Montalbano BG, Mullaney EJ, Dischinger HC, Ullah AHJ (1993) Identification and cloning of a second phytase gene (phyB) from Aspergillus niger (ficcum). Biochem Biophys Res Commun 195:53–57
Ganzhorn AJ, Chanal MC (1990) Kinetic studies with myo-inositol monophosphatase from bovine brain. Biochemistry 29:6065–6071
Geier C, Figura K von, Pohlmann R (1991) Molecular cloning of the mouse lysosomal acid phosphatase. Biol Chem 372:301–304
Gibson D (1987) Production of extracellular phytase from Aspergillus ficuum on starch media. Biotechnol Lett 9:305–310
Gibson DM, Ullah AH (1988) Purification and characterization of phytase from cotyledons of germinating soybean seeds. Arch Biochem Biophys 260:503–513
Gibson DM, Ullah, AB (1990) Phytase and their action on phytic acid in inositol metabolism in plants. Arch Biochem Biophys 262:77–92
Goel M, Sharma CB (1979) Multiple forms of phytase in germinating cotyledons of Cucurbita maxima. Phytochemistry 18:1939–1942
Greaves MP, Anderson G, Webley DM (1967) The hydrolysis of inositol phosphates by Aerobacter aerogenes. Biochim Biophys Acta 132:412–418
Greiner R, Konietzny U, Jany KD (1993) Purification and characterization of two phytases from Escherichia coli. Arch Biochem Biophys 303:107–113
Greiner R, Haller E, Konietzny U, Jany KD (1997) Purification and characterization of a phytase from Klebsiella terrigena. Arch Biochem Biophys 341:201–206
Greiner R, Alminger ML, Carlsson NG (2001) Stereospecificity of myo-inositol hexakisphosphate dephosphorylation by a phytate-degrading enzyme of baker′s yeast. J Agric Food Chem 49:2228–2233
Ha NC, Oh BC, Shin S, Kim HJ, Oh TK, Kim YO, Choi KY, Oh BH (2000) Crystal structures of a novel, thermostable phytase in partially and fully calcium-loaded states. Nat Struct Biol 7:147–153
Hara A, Ebina S, Kondo A, Funagua T (1985) A new type of phytase from Typha latifolia L. Agric Biol Chem 49:3539–3544
Honig B, Nicholls A (1995) Classical electrostatics in biology and chemistry. Science 268:1144–1149
Houde RL, Alli I, Kermasha S (1990) Purification and characterisation of canola seed (Brassica sp.) phytase. J Food Biochem 14:331–351
Idriss EE, Makarewicz O, Farouk A, Rosner K, Greiner R, Bochow H, Richter T, Borriss R (2002) Extracellular phytase activity of Bacillus amyloliquefaciens FZB45 contributes to its plant-growth-promoting effect. Microbiology 148:2097–2109
Irvine GCJ, Cosgrove, DJ (1971) Inositol phosphate phosphatases of microbiological origin. J Biol Sci 24:547
Irving GC, Cosgrove DJ (1972) Inositol phosphate phosphatases of microbiological origin: the inositol pentaphosphate products of Aspergillus ficuum phytases. J Bacteriol 112:434–438
IUPAC–IUB (1975) Enzyme nomenclature recommendation supplement 1: correction and additions. Biochem Biophys Acta 429:1
Jareonkitmongkol S, Ohya M, Watanabe R, Takagi H, Nakamori S (1997) Partial purification of phytase from a soil isolate bacterium, Klebsiella oxytoca MO-3. J Ferment Bioeng 83:393–394
Jonson LF, Tate ME (1969) The conformational analysis of phytic acid based on NMR spectra. J Chem 47:63–73
Kerovuo J, Lauraeus M, Nurminen P, Kalkkinen N, Apajalahti J (1998) Isolation, characterization, molecular gene cloning, and sequencing of a novel phytase from Bacillus subtilis. Appl Environ Microbiol 64:2079–2085
Kerovuo J, Rouvinen J, Hatzack F (2000) Analysis of myo-inositol hexakisphosphate hydrolysis by Bacillus phytase: indication of a novel reaction mechanism. Biochem J 352 Pt 3:623–628
Kim YO, Kim HK, Bae KS, Yu JH, Oh TK (1998a) Purification and properties of a thermostable phytase from Bacillus sp. DS11. Enzyme Microb Technol 22:2–7
Kim YO, Lee JK, Kim HK, Yu JH, Oh TK (1998b) Cloning of the thermostable phytase gene (phy) from Bacillus sp. DS11 and its overexpression in Escherichia coli. FEMS Microbiol Lett 162:185–191
Konietzny U, Greiner R, Jany K-D (1995) Purification and characterisation of a phytase from spelt. J Food Biochem 18:165–183
Kostrewa D, Wyss M, D′Arcy A, Loon AP van (1999) Crystal structure of Aspergillus niger pH 2.5 acid phosphatase at 2.4 Å resolution. J Mol Biol 288:965–974
Kunst F, et al (1997) The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249–256
Laboure AM, Gagnon J, Lescure AM (1993) Purification and characterization of a phytase (myo-inositol- hexakisphosphate phosphohydrolase) accumulated in maize (Zea mays) seedlings during germination. Biochem J 295:413–419
Lassen SF, et al (2001) Expression, gene cloning, and characterization of five novel phytases from four basidiomycete fungi: Peniophora lycii, Agrocybe pediades, a Ceriporia sp., and Trametes pubescens. Appl Environ Microbiol 67:4701–4707
Lim D, Golovan S, Forsberg CW, Jia Z (2000) Crystal structures of Escherichia coli phytase and its complex with phytate. Nat Struct Biol 7:108–113
Lim PE, Tate ME (1973) The phytases. II. Properties of phytase fractions F1 and F2 from wheat bran and the myo-inositol phosphates produced by fraction F2. Biochim Biophys Acta 302:316–328
Lindqvist Y, Schneider G, Vihko P (1994) Crystal structures of rat acid phosphatase complexed with the transition-state analogs vanadate and molybdate. Implications for the reaction mechanism. Eur J Biochem 221:139–142
Liu BL, Rafiq A, Tzeng YM, Rob A (1998) The induction and characterization of phytase and beyond. Enzyme Microb Technol 22:415–424
Maenz DD, Engele-Schaan CM, Newkirk RW, Classen HL (1999) The effect of minerals and mineral chelators on the formation of phytase-resistant and phytase-susceptible forms of phytic acid in solution and in a slurry of canola meal. Anim Feed Sci Technol 81:177–192
Maiti IB, Majumder, AL, Biswas BB (1974) Purification and mode of action of phytase from Phaseolus aureus. Phytochemistry 13:1047–1051
Mandal NC, Burnman S, Biswas, BB (1972) Isolation, purification and characterization of phytase from germinating mung beans. Phytochemistry 11:495–502
Martin CJ, Evans WJ (1986) Phytic acid–metal ion interactions II. The effect of pH on Ca(II) binding. J Inorg Biochem 27:17–30
Meier B, Scherk C, Schmidt M, Parak F (1998) pH-dependent inhibition by azide and fluoride of the iron superoxide dismutase from Propionibacterium shermanii. Biochem J 331:403–407
Mitchell DB, Vogel K, Weimann BJ, Pasamontes L, Loon AP van (1997) The phytase subfamily of histidine acid phosphatases: isolation of genes for two novel phytases from the fungi Aspergillus terreus and Myceliophthora thermophila. Microbiology 143:245–252
Nagai M, Nishibu M, Sugita Y, Yoneyama Y (1975) The effects of inositol hexaphosphate on the allosteric properties of two beta-99-substituted abnormal hemoglobins, hemoglobin Yakima and hemoglobin Kempsey. J Biol Chem 250:3169–3173
Nayini NRP (1984) The phytase of yeast. Lebensm Wiss Technol 17:24–26
Nelson TS, Shieh TR, Wodzinski RJ, Ware JH (1968) The availability of phytate phosphorus in soybean meal before and after treatment with a mold phytase. Poult Sci 47:1842–1848
Oh BC, Chang BS, Park KH, Ha NC, Kim HK, Oh BH, Oh TK (2001) Calcium-dependent catalytic activity of a novel phytase from Bacillus amyloliquefaciens DS11. Biochemistry 40:9669–9676
Ostanin K, Van Etten RL (1993) Asp304 of Escherichia coli acid phosphatase is involved in leaving group protonation. J Biol Chem 268:20778–20784
Ostanin K, Harms EH, Stevis PE, Kuciel R, Zhou MM, Van Etten RL (1992) Overexpression, site-directed mutagenesis, and mechanism of Escherichia coli acid phosphatase. J Biol Chem 267:22830–22836
Pasamontes L, Haiker M, Henriquez-Huecas M, Mitchell DB, Loon AP van (1997a) Cloning of the phytases from Emericella nidulans and the thermophilic fungus Talaromyces thermophilus. Biochim Biophys Acta 1353:217–223
Pasamontes L, Haiker M, Wyss M, Tessier M, Loon AP van (1997b) Gene cloning, purification, and characterization of a heat-stable phytase from the fungus Aspergillus fumigatus. Appl Environ Microbiol 63:1696–1700
Piddington CS, et al (1993) The cloning and sequencing of the genes encoding phytase (phy) and pH 2.5 optimum acid phosphatase (aph) from Aspergillus niger var awamori. Gene 133:55–62
Pohlmann R, Houston CS, Paloheimo M, Cantrell M, Miettinen-Oinonen A, Nevalainen H, Rambosek J (1988) Human lysosomal acid phosphatase: cloning, expression and chromosomal assignment. EMBO J 7:2343–2350
Porvari KS, Herrala AM, Kurkela RM, Taavitsainen PA, Lindqvist Y, Schneider G, Vihko PT (1994) Site-directed mutagenesis of prostatic acid phosphatase. Catalytically important aspartic acid 258, substrate specificity, and oligomerization. J Biol Chem 269:22642–22646
Powar VK, Jagannathan V (1982) Purification and properties of phytate-specific phosphatase from Bacillus subtilis. J Bacteriol 151:1102–1108
Reddy NR, Sathe SK, Salunkhe DK (1982) Phytates in legumes and cereals. Adv Food Res 28:1–92
Roiko K, Janne OA, Vihko P (1990) Primary structure of rat secretory acid phosphatase and comparison to other acid phosphatases. Gene 89:223–229
Schneider G, Lindqvist Y, Vihko P (1993) Three-dimensional structure of rat acid phosphatase. EMBO J 12:2609–2615
Schroder B, Breves G, Rodehutscord M (1996) Mechanisms of intestinal phosphorus absorption and availability of dietary phosphorus in pigs. Dtsch Tieraerztl Wochenschr 103:209–214
Scott JJ (1991) Alkaline phytase activity in nonionic detergent extracts of legume seeds. Plant Physiol 95:1298–1301
Scott JJ, Loewus FA (1986) A calcium-activated phytase from pollen of Lilium longiflorum. Plant Physiol 82:333–335
Segel IH (1975) In: Segel IH (ed) Enzyme kinetics: substrate–activator complex is the true substrate. Wiley, New York, pp 242–272
Segueilha L, Lambrechts C, Boze H, Moulin G, Galzy P (1992) Purification and properties of the phytase from Schwanniomyces castellii. J Ferment Bioeng 74:7–11
Shah V, Parekh LJ (1990) Phytase from Klebsiella sp. No. PG-2: purification and properties. Indian J Biochem Biophys 27:98–102
Shieh TR, Ware JH (1968) Survey of microorganisms for the production of extracellular phytase. Appl Microbiol 16:1348–1351
Shimizu M (1992) Purification and characterization of phytase from Bacillus subtilis (natto) N-77. Biosci Biotechnol Biochem 56:1266–1269
Shimizu M (1993) Purification and characterization of phytase and acid phosphatase produced by Aspergillus oryzae K1. Biosci Biotechnol Biochem 57:1364–1365
Shin S, Ha NC, Oh BC, Oh TK, Oh BH (2001) Enzyme mechanism and catalytic property of beta propeller phytase. Structure 9:851–858
Skowronski T (1978) Some properties of partially purified phytase from Aspergillus niger. Acta Microbiol Pol 27:41–48
Sutardi, Buckle KA (1986) The characteristics of soybean phytase. J Food Biochem 10:197–216
Tambe SM, Kaklij GS, Kelkar SM, Parekh LJ (1994) Two distinct molecular forms of phytase from Klebsiella aerogenes: evidence for unusually small active enzyme peptide. J Ferment Bioeng 77:23–27
Tomschy A, et al (2002) Engineering of phytase for improved activity at low pH. Appl Environ Microbiol 68:1907–1913
Tye AJ, Siu FK, Leung TY, Lim BL (2002) Molecular cloning and the biochemical characterization of two novel phytases from B. subtilis 168 and B. licheniformis. Appl Microbiol Biotechnol 59:190–197
Ullah AH (1988) Aspergillus ficuum phytase: partial primary structure, substrate selectivity, and kinetic characterization. Prep Biochem 18:459–471
Ullah AH, Cummins BJ (1987) Purification, N-terminal amino acid sequence and characterization of pH 2.5 optimum acid phosphatase (EC 3.1.3.2) from Aspergillus ficuum. Prep Biochem 17:397–422
Ullah AH, Phillippy BQ (1994) Substrate selectivity in Aspergillus ficcum phytase and acid phosphtase using myo-inositol phosphates. J Agric Food Chem 42:423–425
Ullah AH, Sethumadhavan K, Lei XG, Mullaney EJ (2000) Biochemical characterization of cloned Aspergillus fumigatus phytase (phyA). Biochem Biophys Res Commun 275:279–285
Urbano G, Lopez-Jurado M, Aranda P, Vidal-Valverde C, Tenorio E, Porres J (2000) The role of phytic acid in legumes: antinutrient or beneficial function? J Physiol Biochem 56:283–294
Van der Klis JD, Versteegh HA, Simons PC, Kies AK (1997) The efficacy of phytase in corn-soybean meal-based diets for laying hens. Poult Sci 76:1535–1542
Van Etten RL, Davidson R, Stevis PE, MacArthur H, Moore DL (1991) Covalent structure, disulfide bonding, and identification of reactive surface and active site residues of human prostatic acid phosphatase. J Biol Chem 266:2313–2319
Van Hartingsveldt W, et al (1993) Cloning, characterization and overexpression of the phytase-encoding gene (phyA) of Aspergillus niger. Gene 127:87–94
Vincent JB, Crowder MW, Averill BA (1992) Hydrolysis of phosphate monoesters: a biological problem with multiple chemical solutions. Trends Biochem Sci 17:105–110
Wyss M, Brugger R, Kronenberger A, Remy R, Fimbel R, Oesterhelt G, Lehmann M, Loon AP van (1999a) Biochemical characterization of fungal phytases (myo-inositol hexakisphosphate phosphohydrolases): catalytic properties. Appl Environ Microbiol 65:367–373
Wyss M, Pasamontes L, Friedlein A, Remy R, Tessier M, Kronenberger A, Middendorf A, Lehmann M, Schnoebelen L, Rothlisberger U, Kusznir E, Wahl G, Muller F, Lahm HW, Vogel K, Loon AP van (1999b) Biophysical characterization of fungal phytases (myo-inositol hexakisphosphate phosphohydrolases): molecular size, glycosylation pattern, and engineering of proteolytic resistance. Appl Environ Microbiol 65:359–366
Yamada K, Minoda, Y, Yamada K (1968) Phytase from Aspergillus terreus. Part 1. Production, purification, and some general properties of the enzyme. Agric Biol Chem 32:1275–1283
Yamamoto S, Minoda Y, Yamada K (1972) Chemical and physicochemical properties of phytase from Aspergillus terreus. Agric Biol Chem 36:2097–2103
Acknowledgements.
The authors gratefully acknowledge Drs. M. Rudolph, S-H Lee, and J-W Kim for critical reading of the manuscript. This study was supported by the G7 project and in part by the 21C Frontier R&D Program from the Korean Ministry of Science and Technology.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Oh, BC., Choi, WC., Park, S. et al. Biochemical properties and substrate specificities of alkaline and histidine acid phytases. Appl Microbiol Biotechnol 63, 362–372 (2004). https://doi.org/10.1007/s00253-003-1345-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-003-1345-0