Abstract
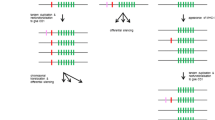
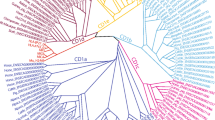
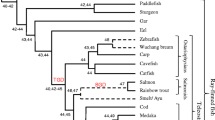
CD1, as the third family of antigen-presenting molecules, is previously only found in mammals and chickens, which suggests that the chicken and mammalian CD1 shared a common ancestral gene emerging at least 310 million years ago. Here, we describe CD1 genes in the green anole lizard and Crocodylia, demonstrating that CD1 is ubiquitous in mammals, birds, and reptiles. Although the reptilian CD1 protein structures are predicted to be similar to human CD1d and chicken CD1.1, CD1 isotypes are not found to be orthologous between mammals, birds, and reptiles according to phylogenetic analyses, suggesting an independent diversification of CD1 isotypes during the speciation of mammals, birds, and reptiles. In the green anole lizard, although the single CD1 locus and MHC I gene are located on the same chromosome, there is an approximately 10-Mb-long sequence in between, and interestingly, several genes flanking the CD1 locus belong to the MHC paralogous region on human chromosome 19. The CD1 genes in Crocodylia are located in two loci, respectively linked to the MHC region and MHC paralogous region (corresponding to the MHC paralogous region on chromosome 19). These results provide new insights for studying the origin and evolution of CD1.







Similar content being viewed by others
References
Adams EJ, Luoma AM (2013) The adaptable major histocompatibility complex (MHC) fold: structure and function of nonclassical and MHC class I-like molecules. Annu Rev Immunol 31:529–561. doi:10.1146/annurev-immunol-032712-095912
Albertson DG, Fishpool R, Sherrington P, Nacheva E, Milstein C (1988) Sensitive and high resolution in situ hybridization to human chromosomes using biotin labelled probes: assignment of the human thymocyte CD1 antigen genes to chromosome 1. EMBO J 7:2801–2805
Balk SP, Ebert EC, Blumenthal RL, McDermott FV, Wucherpfennig KW, Landau SB, Blumberg RS (1991) Oligoclonal expansion and CD1 recognition by human intestinal intraepithelial lymphocytes. Science 253:1411–1415
Blumberg RS, Gerdes D, Chott A, Porcelli SA, Balk SP (1995) Structure and function of the CD1 family of MHC-like cell surface proteins. Immunol Rev 147:5–29
Brigl M, Brenner MB (2004) CD1: antigen presentation and T cell function. Annu Rev Immunol 22:817–890. doi:10.1146/annurev.immunol.22.012703.104608
Calabi F, Milstein C (1986) A novel family of human major histocompatibility complex-related genes not mapping to chromosome 6. Nature 323:540–543. doi:10.1038/323540a0
Castano AR (1995) Peptide binding and presentation by mouse CD1. Science 269:223–226
Dascher CC (2007) Evolutionary biology of CD1. Curr Top Microbiol Immunol 314:3–26
Dascher CC, Brenner MB (2003) Evolutionary constraints on CD1 structure: insights from comparative genomic analysis. Trends Immunol 24:412–418
Dougan SK, Kaser A, Blumberg RS (2007) CD1 expression on antigen-presenting cells. Curr Top Microbiol Immunol 314:113–141
Hein J (1990) [39] Unified approach to alignment and phylogenies. In: Methods in Enzymology, vol Volume 183. Academic Press, pp 626–645. doi:http://dx.doi.org/10.1016/0076-6879(90)83041-7
Hokamp K, McLysaght A, Wolfe KH (2003) The 2R hypothesis and the human genome sequence. J Struct Funct Genom 3:95–110
Holland PW, Garcia-Fernandez J, Williams NA, Sidow A (1994) Gene duplications and the origins of vertebrate development Development (Cambridge, England) Supplement:125–133
Jayawardena-Wolf J, Bendelac A (2001) CD1 and lipid antigens: intracellular pathways for antigen presentation. Curr Opin Immunol 13:109–113
Kasahara M (1999) The chromosomal duplication model of the major histocompatibility complex. Immunol Rev 167:17–32
Klein J et al (1990) Nomenclature for the major histocompatibility complexes of different species: a proposal. Immunogenetics 31:217–219
Koch M et al (2005) The crystal structure of human CD1d with and without alpha-galactosylceramide. Nat Immunol 6:819–826
Kumar S, Hedges SB (1998) A molecular timescale for vertebrate evolution. Nature 392:917–920. doi:10.1038/31927
Martin LH, Calabi F, Milstein C (1986) Isolation of CD1 genes: a family of major histocompatibility complex-related differentiation antigens. Proc Natl Acad Sci U S A 83:9154–9158
Maruoka T, Tanabe H, Chiba M, Kasahara M (2005) Chicken CD1 genes are located in the MHC: CD1 and endothelial protein C receptor genes constitute a distinct subfamily of class-I-like genes that predates the emergence of mammals. Immunogenetics 57:590–600. doi:10.1007/s00251-005-0016-y
Matsuda JL, Kronenberg M (2001) Presentation of self and microbial lipids by CD1 molecules. Curr Opin Immunol 13:19–25
McMichael AJ, Pilch JR, Galfre G, Mason DY, Fabre JW, Milstein C (1979) A human thymocyte antigen defined by a hybrid myeloma monoclonal antibody. Eur J Immunol 9:205–210
Miller MM et al (2005) Characterization of two avian MHC-like genes reveals an ancient origin of the CD1 family. Proc Natl Acad Sci U S A 102:8674–8679. doi:10.1073/pnas.0500105102
Moody DB et al (2004) T cell activation by lipopeptide antigens. Science 303:527–531. doi:10.1126/science.1089353
Ohno S (1970) Evolution by gene duplication. George Alien & Unwin Ltd. Berlin, Heidelberg and New York: Springer-Verlag, London
Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Applic Biosci : CABIOS 12:357–358
Porcelli SA (1995) The CD1 family: a third lineage of antigen-presenting molecules. Adv Immunol 59:1–98
Porcelli SA, Modlin RL (1999) The CD1 system: antigen-presenting molecules for T cell recognition of lipids and glycolipids. Annu Rev Immunol 17:297–329. doi:10.1146/annurev.immunol.17.1.297
Porcelli S, Brenner MB, Greenstein JL, Balk SP, Terhorst C, Bleicher PA (1989) Recognition of cluster of differentiation 1 antigens by human CD4-CD8-cytolytic T lymphocytes. Nature 341:447–450
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Salomonsen J et al (2005) Two CD1 genes map to the chicken MHC, indicating that CD1 genes are ancient and likely to have been present in the primordial MHC. Proc Natl Acad Sci U S A 102:8668–8673. doi:10.1073/pnas.0409213102
Sandoval IV, Bakke O (1994) Targeting of membrane proteins to endosomes and lysosomes. Trends Cell Biol 4:292–297
Sugita M et al (1996) Cytoplasmic tail-dependent localization of CD1b antigen-presenting molecules to MIICs. Science 273:349–352
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Wan QH et al (2013) Genome analysis and signature discovery for diving and sensory properties of the endangered Chinese alligator. Cell Res 23:1091–1105
Zajonc DM, Striegl H, Dascher CC, Wilson IA (2008) The crystal structure of avian CD1 reveals a smaller, more primordial antigen-binding pocket compared to mammalian CD1. Proc Natl Acad Sci U S A 105:17925–17930
Acknowledgments
This study was supported by the National Natural Science Foundation of China (31272433).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 1802 kb)
Rights and permissions
About this article
Cite this article
Yang, Z., Wang, C., Wang, T. et al. Analysis of the reptile CD1 genes: evolutionary implications. Immunogenetics 67, 337–346 (2015). https://doi.org/10.1007/s00251-015-0837-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-015-0837-2