Abstract
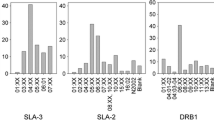
In order to identify and characterize genetic polymorphism of the swine major histocompatibility complex (Mhc: SLA) class I genes, RT-PCR products of the second and third exons of the three SLA classical class I genes, SLA-1, SLA-2 and SLA-3 were subjected to nucleotide determination. These analyses allowed the identification of four, eight and seven alleles at the SLA-1, SLA-2 and SLA-3 loci, respectively, from three different breeds of miniature swine and one mixed breed. Among them, 12 alleles were novel. Construction of a phylogenetic tree using the nucleotide sequences of those 19 alleles indicated that the SLA-1 and -2 genes are more closely related to each other than to SLA-3. Selective forces operating at single amino acid sites of the SLA class I molecules were analyzed by the Adaptsite Package program. Ten positive selection sites were found at the putative antigen recognition sites (ARSs). Among the 14 positively selected sites observed in the human MHC (HLA) classical class I molecules, eight corresponding positions in the SLA class I molecules were inferred as positively selected. On the other hand, four amino acids at the putative ARSs were identified as negatively selected in the SLA class I molecules. These results suggest that selective forces operating in the SLA class I molecules are almost similar to those of the HLA class I molecules, although several functional sites for antigen and cytotoxic T-lymphocyte recognition by the SLA class I molecules may be different from those of the HLA class I molecules.




Similar content being viewed by others
References
Blangero J, Tissot RG, Beattie CW, Amoss MS Jr (1996) Genetic determinants of cutaneous malignant melanoma in Sinclair swine. Br J Cancer 73:667–671
Bjorkman PJ, Saper MA, Samrauoi B, Bennett WS, Strominger JL, Wiley DC (1987) The foreign antigen binding site and T cell recognition regions of class I histocompatibility complex. Nature 329:512–518
Chardon P (1999) The major histocompatibility complex in swine. Immunol Rev 167:179–192
Chardon P, Rogel-Gaillard C, Cattolico L, Duprat S, Vaiman M, Renard C (2001) Sequence of the pig major histocompatibility region containing all non-classical class I genes. Tissue Antigens 57:55–65
Conley AJ, Jung YC, Schwartz NK, Warner CM, Rothschild MF, Ford SP (1988) Influence of SLA haplotypes on ovulation rate and litter size in miniature pigs. J Reprod Fertil 82:595–601
Ehlich R, Lifshitz R, Pescovitz MD, Rudikoff S, Singer DS (1987) Tissue-specific expression and structure of a divergent member of a class I MHC gene family. J Immunol 139:593–602
Gu X, Nei M (1999) Locus specificity of polymorphic alleles and evolution by a birth-and-death process in mammalian MHC genes. Mol Biol Evol 16:147–156
Kumar S, Tamura K, Jakobsen IB, Nei M (2001) MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244–1245
Nakanishi Y, Ogawa K, Yanagita K, Yamauchi C (1991) Body measurement and some characteristics of inbred Clawn miniature pigs. Jpn J Swine Sci 28:211–218
Parham P, Lomen CE, Lawlor DA, Ways P, Holmes N, Coppin HL, Salter RD, Wan AM, Ennis PD (1988) Nature of polymorphism in HLA-A, -B and -C molecules. Proc Natl Acad Sci USA 85:4005–4008
Renard C, Viman M, Capy P, Sellier P (1982) SLA markers and characters of production in the pig. Second World Congress on Genetics Applied to Livestock Production, vol 8, pp 570–583
Renard C, Viman M, Chiannilkuchi N, Cattolico L, Robert C, Chardon P (2001) Sequence of the pig major histocompatibility region containing the classical class I genes. Immunogenetics 53:490–500
Renard C, Chardon P, Viman M (2003) The phylogenetic history of the MHC class I gene families in pig, including a fossil gene predating mammalian radiation. J Mol Evol (in press)
Saitou N (1989) A theoretical study of the underestimation of branch lengths by the maximum parsimony principle. Syst Zool 38:1-6
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Salter RD, Benjamin RJ, Wesley PK, Buxton SE, Garrett TPJ, Clayberger C, Kensky AM, Nomment AM, Littman DR, Parham P (1990) A binding site for the T cell co-receptor CD8 on the alpha-3 domain of HLA-A2. Nature 345:41–46
Satz ML, Wang L-C, Singer DS, Rudikoff S (1985) Structure and expression of two porcine genomic clones encoding class I MHC antigens. J Immunol 135:2167–2175
Shishido S, Naziruddin B, Howard T, Mohanakumar T (1997) Recognition of porcine major histocompatibility complex class I antigens by human CD8+ cytolytic T cell clones. Transplantation 64:340–346
Singer DS, Ehrlich R, Golding H, Satz ML, Parent L, Rudikoff S (1987) Structure and expression of class I MHC genes in the miniature swine. In: Warner C, Rothschild M, Lamont S (eds) Molecular biology of the major histocompatibility complex of domestic animal species. Iowa State University Press, Ames, pp 53–62
Sullivan JA, Oettinger HF, Sachs DH, Edge AS (1997) Analysis of polymorphism in porcine Mhc class I genes. J Immunol 159:2318–2326
Suzuki Y, Gojobori T (1999) A method for detecting positive selection at single amino acid sites. Mol Biol Evol 16:1315–1328
Suzuki Y, Gojobori T, Nei (2001) ADAPTSITE: detecting natural selection at single amino acid sites. Bioinformatics 17:660–661
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions is the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Yamada K, Sachs DH, Der Simonian H (1995) Human anti-porcine xenogenic T cell response: evidence for allelic specificity of mixed leukocyte reaction and for both direct and indirect pathway of recognition. J Immunol 155:5249–5256
Velten F, Rogel-Gaillard, C, Renald C, Pontarotti, P, Tazi-Ahnini R, Vaiman M, Chardon P (1998) A first map of the porcine major histocompatibility complex class I region. Tissue Antigens 51:183–194
Velten F, Renald C, Rogel-Gaillard C, Vaiman M, Schrezenmeir J, Chardon P (1999) Spatial arrangement of pig Mhc class I sequences. Immunogenetics 49:919–930
Wu TT, Kabat E (1970) An analysis of the sequences of the variable of Bence Jones proteins and myeloma light chains and their implications for antibody complementarity. J Exp Med 132:211–250
Acknowledgements
We would like to thank Dr. Patrick Chardon (INRA-CEA, France) for useful discussion. We also wish to thank Dr. Makiko Shiba and Mariko Tsustui (Ibaraki Station, National Livestock Breeding Center, Independent Administrative Institution) for providing the peripheral blood samples from Mexican hairless pigs, and Mr. Daisuke Sumiyama (Tokai University School of Medicine) for technical assistance. This study was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and the Animal Genome Research Project of the Ministry of Agriculture, Forestry and Fisheries of Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
The DNA sequence data reported in this paper have been submitted to the DDBJ, EMBL and GenBank nucleotide databases and have been assigned the accession numbers, AB105379, AB105380, AB105381, AB105382, AB105383, AB105384, AB105385, AB105386, AB105388, AB105389, AB105390 and AB105391
Rights and permissions
About this article
Cite this article
Ando, A., Kawata, H., Shigenari, A. et al. Genetic polymorphism of the swine major histocompatibility complex (SLA) class I genes, SLA-1, -2 and -3 . Immunogenetics 55, 583–593 (2003). https://doi.org/10.1007/s00251-003-0619-0
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-003-0619-0