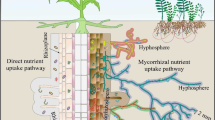
Abstract
Rhizosphere microbes influence one another, forming extremely complex webs of interactions that may determine plant success. Identifying the key factors that structure the fungal microbiome of the plant rhizosphere is a necessary step in optimizing plant production. In a long-term field experiment conducted at three locations in the Canadian prairies, we tested the following hypotheses: (1) diversification of cropping systems influences the fungal microbiome of the canola (Brassica napus) rhizosphere; (2) the canola rhizosphere has a core fungal microbiome, i.e., a set of fungi always associated with canola; and (3) some taxa within the rhizosphere microbiome of canola are highly interrelated and fit the description of hub taxa. Our results show that crop diversification has a significant effect on the structure of the rhizosphere fungal community but not on fungal diversity. We also discovered and described a canola core microbiome made up of one zero-radius operational taxonomic unit (ZOTU), cf. Olpidium brassicae, and an eco-microbiome found only in 2013 consisting of 47 ZOTUs. Using network analysis, we identified four hub taxa in 2013: ZOTU14 (Acremonium sp.), ZOTU28 (Sordariomycetes sp.), ZOTU45 (Mortierella sp.) and ZOTU179 (cf. Ganoderma applanatum), and one hub taxon, ZOTU17 (cf. Mortierella gamsii) in 2016. None of these most interacting taxa belonged to the core microbiome or eco-microbiome for each year of sampling. This temporal variability puts into question the idea of a plant core fungal microbiome and its stability. Our results provide a basis for the development of ecological engineering strategies for the improvement of canola production systems in Canada.



Similar content being viewed by others
References
Bais HP, Weir TL, Perry LG et al (2006) The role of root exudates in rhizosphere interactions with plants and other organisms. Annual review of plant biology. Annual Reviews, Palo Alto, pp 233–266
Tkacz A, Cheema J, Chandra G, Grant A, Poole PS (2015) Stability and succession of the rhizosphere microbiota depends upon plant type and soil composition. ISME J 9:2349–2359. https://doi.org/10.1038/ismej.2015.41
Pii Y, Borruso L, Brusetti L, Crecchio C, Cesco S, Mimmo T (2016) The interaction between iron nutrition, plant species and soil type shapes the rhizosphere microbiome. Plant Physiol Biochem 99:39–48. https://doi.org/10.1016/j.plaphy.2015.12.002
Garbaye J (1994) Helper bacteria - a new dimension to the mycorrhizal symbiosis. New Phytol 128:197–210. https://doi.org/10.1111/j.1469-8137.1994.tb04003.x
van der Heijden MG, Streitwolf-Engel R, Riedl R, Siegrist S, Neudecker A, Ineichen K, Boller T, Wiemken A, Sanders IR (2006) The mycorrhizal contribution to plant productivity, plant nutrition and soil structure in experimental grassland. New Phytol 172:739–752. https://doi.org/10.1111/j.1469-8137.2006.01862.x
Doehlemann G, Requena N, Schaefer P, Brunner F, O’Connell R, Parker JE (2014) Reprogramming of plant cells by filamentous plant-colonizing microbes. New Phytol 204:803–814. https://doi.org/10.1111/nph.12938
Hardoim PR, van Overbeek LS, van Elsas JD (2008) Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol 16:463–471. https://doi.org/10.1016/j.tim.2008.07.008
Latz E, Eisenhauer N, Rall BC, Scheu S, Jousset A (2016) Unravelling linkages between plant community composition and the pathogen-suppressive potential of soils. Sci Rep 6:23584. https://doi.org/10.1038/srep23584
Mozafar A, Anken T, Ruh R, Frossard E (2000) Tillage intensity, mycorrhizal and nonmycorrhizal fungi, and nutrient concentrations in maize, wheat, and canola. Agron J 92:1117. https://doi.org/10.2134/agronj2000.9261117x
Bulgarelli D, Schlaeppi K, Spaepen S, ver Loren van Themaat E, Schulze-Lefert P (2013) Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64:807–838. https://doi.org/10.1146/annurev-arplant-050312-120106
van der Heijden MG, de Bruin S, Luckerhoff L, van Logtestijn R, Schlaeppi K (2016) A widespread plant-fungal-bacterial symbiosis promotes plant biodiversity, plant nutrition and seedling recruitment. ISME J 10:389–399. https://doi.org/10.1038/ismej.2015.120
Vandenkoornhuyse P, Quaiser A, Duhamel M, le van A, Dufresne A (2015) The importance of the microbiome of the plant holobiont. New Phytol 206:1196–1206. https://doi.org/10.1111/nph.13312
Rout ME (2014). Plant Microbiome 69:279–309. https://doi.org/10.1016/b978-0-12-417163-3.00011-1
Agler MT, Ruhe J, Kroll S, Morhenn C, Kim ST, Weigel D, Kemen EM (2016) Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol 14:e1002352. https://doi.org/10.1371/journal.pbio.1002352
Taktek S, St-Arnaud M, Piché Y, Fortin JA, Antoun H (2017) Igneous phosphate rock solubilization by biofilm-forming mycorrhizobacteria and hyphobacteria associated with Rhizoglomus irregulare DAOM 197198. Mycorrhiza 27:13–22. https://doi.org/10.1007/s00572-016-0726-z
Hajishengallis G, Darveau RP, Curtis MA (2012) The keystone-pathogen hypothesis. Nat Rev Microbiol 10:717–725. https://doi.org/10.1038/nrmicro2873
Raaijmakers JM, Paulitz TC, Steinberg C et al (2009) The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321:341–361. https://doi.org/10.1007/s11104-008-9568-6
Lay C-Y, Bell TH, Hamel C et al (2018) Canola root–associated microbiomes in the canadian prairies. Front Microbiol:9. https://doi.org/10.3389/fmicb.2018.01188
Zheng ZT, Pei FC, Jiang LP, Fu CQ (2014) Synthesis and characterization of canola oil-based polyisocyanates. In: Adv Mater Res. https://www.scientific.net/AMR.955-959.802. Accessed 15 Jul 2018
Rumberger A, Marschner P (2003) 2-Phenylethylisothiocyanate concentration and microbial community composition in the rhizosphere of canola. Soil Biol Biochem 35:445–452. https://doi.org/10.1016/s0038-0717(02)00296-1
Harker KN, O’Donovan JT, Turkington TK et al (2015) Canola rotation frequency impacts canola yield and associated pest species. Can J Plant Sci 95:9–20. https://doi.org/10.4141/cjps-2014-289
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. https://doi.org/10.1093/bioinformatics/btq461
Dorn-In S, Hölzel CS, Janke T et al (2013) PCR-SSCP-based reconstruction of the original fungal flora of heat-processed meat products. Int J Food Microbiol 162:71–81. https://doi.org/10.1016/j.ijfoodmicro.2012.12.022
McCune B, Mefford MJ (2011) PC-ORD multivariate analysis of ecological data. Version 6
Kurtz ZD, Müller CL, Miraldi ER, Littman DR, Blaser MJ, Bonneau RA (2015) Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput Biol 11:e1004226. https://doi.org/10.1371/journal.pcbi.1004226
Teakle DS (1960) Association of Olpidium brassicae and tobacco necrosis virus. Nature 188:431–432. https://doi.org/10.1038/188431a0
Campbell RN, Sim ST (1994) Host specificity and nomenclature of Olpidium bornovanus (= Olpidium radicale) and comparisons to Olpidium brassicae. Can J Bot 72:1136–1143
Campbell RN, Sim ST, Lecoq H (1995) Virus transmission by host-specific strains of Olpidium bornovanus and Olpidium brassicae. Eur J Plant Pathol 101:273–282. https://doi.org/10.1007/BF01874783
Sekimoto S, Rochon D, Long JE et al (2011) A multigene phylogeny of Olpidium and its implications for early fungal evolution. BMC Evol Biol 11:331
Hilton S, Bennett AJ, Keane G et al (2013) Impact of shortened crop rotation of oilseed rape on soil and rhizosphere microbial diversity in relation to yield decline. PLoS ONE:8. https://doi.org/10.1371/journal.pone.0059859
Lay C-Y, Hamel C, St-Arnaud M (2018) Taxonomy and pathogenicity of Olpidium brassicae and its allied species. Fungal Biol 122:837–846. https://doi.org/10.1016/j.funbio.2018.04.012
Gaiero JR, McCall CA, Thompson KA, Day NJ, Best AS, Dunfield KE (2013) Inside the root microbiome: bacterial root endophytes and plant growth promotion. Am J Bot 100:1738–1750. https://doi.org/10.3732/ajb.1200572
Edwards J, Johnson C, Santos-Medellin C et al (2015) Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci U S A 112:E911–E920. https://doi.org/10.1073/pnas.1414592112
Coleman-Derr D, Desgarennes D, Fonseca-Garcia C, Gross S, Clingenpeel S, Woyke T, North G, Visel A, Partida-Martinez LP, Tringe SG (2016) Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol 209:798–811. https://doi.org/10.1111/nph.13697
Siciliano SD, Theoret CM, de Freitas JR et al (1998) Differences in the microbial communities associated with the roots of different cultivars of canola and wheat. Can J Microbiol 44:844–851. https://doi.org/10.1139/w98-075
Gan Y, Hamel C, O’Donovan JT et al (2015) Diversifying crop rotations with pulses enhances system productivity. Sci Rep 5:14625. https://doi.org/10.1038/srep14625
Smith EG, Kutcher HR, Brandt SA et al (2013) The profitability of short-duration canola and pea rotations in Western Canada. Can J Plant Sci 93:933–940. https://doi.org/10.4141/cjps2013-021
Larkin RP (2003) Characterization of soil microbial communities under different potato cropping systems by microbial population dynamics, substrate utilization, and fatty acid profiles. Soil Biol Biochem 35:1451–1466. https://doi.org/10.1016/s0038-0717(03)00240-2
Larkin RP, Honeycutt CW (2006) Effects of different 3-year cropping systems on soil microbial communities and rhizoctonia diseases of potato. Phytopathology 96:68–79. https://doi.org/10.1094/PHYTO-96-0068
Bennett AJ, Hilton S, Bending GD, Chandler D, Mills P (2014) Impact of fresh root material and mature crop residues of oilseed rape (Brassica napus) on microbial communities associated with subsequent oilseed rape. Biol Fertil Soils 50:1267–1279. https://doi.org/10.1007/s00374-014-0934-7
Gkarmiri K, Mahmood S, Ekblad A et al (2017) Identifying the active microbiome associated with roots and rhizosphere soil of oilseed rape. Appl Environ Microbiol 83:e01938–e01917. https://doi.org/10.1128/AEM.01938-17
Schmitt A, Glaser B (2011) Organic matter dynamics in a temperate forest soil following enhanced drying. Soil Biol Biochem 43:478–489. https://doi.org/10.1016/j.soilbio.2010.09.037
Wang N, Wang M, Li S et al (2015) Effects of precipitation variation on growing seasonal dynamics of soil microbial biomass in broadleaved Korean pine mixed forest. Ying Yong Sheng Tai Xue Bao J Appl Ecol 26:1297–1305
Zhang F-G, Zhang Q-G (2016) Microbial diversity limits soil heterotrophic respiration and mitigates the respiration response to moisture increase. Soil Biol Biochem 98:180–185. https://doi.org/10.1016/j.soilbio.2016.04.017
Ochoa-Hueso R, Collins SL, Delgado-Baquerizo M et al (2018) Drought consistently alters the composition of soil fungal and bacterial communities in grasslands from two continents. Glob Chang Biol 24:2818–2827. https://doi.org/10.1111/gcb.14113
James TY, Letcher PM, Longcore JE, Mozley-Standridge SE, Porter D, Powell MJ, Griffith GW, Vilgalys R (2006) A molecular phylogeny of the flagellated fungi (Chytridiomycota) and description of a new phylum (Blastocladiomycota). Mycologia 98:860–871. https://doi.org/10.1080/15572536.2006.11832616
Toberman H, Freeman C, Evans C, Fenner N, Artz RR (2008) Summer drought decreases soil fungal diversity and associated phenol oxidase activity in upland Calluna heathland soil. FEMS Microbiol Ecol 66:426–436. https://doi.org/10.1111/j.1574-6941.2008.00560.x
Florinsky IV, Eilers RG, Manning GR, Fuller LG (2002) Prediction of soil properties by digital terrain modelling. Environ Model Softw 17:295–311. https://doi.org/10.1016/S1364-8152(01)00067-6
de Campos SB, Youn JW, Farina R, Jaenicke S, Jünemann S, Szczepanowski R, Beneduzi A, Vargas LK, Goesmann A, Wendisch VF, Passaglia LM (2013) Changes in root bacterial communities associated to two different development stages of canola (Brassica napus L. var oleifera) evaluated through next-generation sequencing technology. Microb Ecol 65:593–601. https://doi.org/10.1007/s00248-012-0132-9
Wagner MR, Lundberg DS, Coleman-Derr D et al (2015) Corrigendum to Wagneret al.: natural soil microbes alter flowering phenology and the intensity of selection on flowering time in a wild Arabidopsis relative. Ecol Lett 18:218–220. https://doi.org/10.1111/ele.12400
Barnes CJ, van der Gast CJ, Burns CA et al (2016) Temporally variable geographical distance effects contribute to the assembly of root-associated fungal communities. Front Microbiol:7. https://doi.org/10.3389/fmicb.2016.00195
Fernandez MR (2007) Fusarium populations in roots of oilseed and pulse crops grown in eastern Saskatchewan. Can J Plant Sci 87:945–952
Fernandez M, Huber D, Basnyat P, Zentner R (2008) Impact of agronomic practices on populations of Fusarium and other fungi in cereal and noncereal crop residues on the Canadian prairies. Soil Tillage Res 100:60–71. https://doi.org/10.1016/j.still.2008.04.008
Hamel C, Vujanovic V, Jeannotte R et al (2005) Negative feedback on a perennial crop: fusarium crown and root rot of asparagus is related to changes in soil microbial community structure. Plant Soil 268:75–87. https://doi.org/10.1007/s11104-004-0228-1
Vujanovic V, Hamel C, Yergeau E, St-Arnaud M (2006) Biodiversity and biogeography of fusarium species from northeastern North American asparagus fields based on microbiological and molecular approaches. Microb Ecol 51:242–255. https://doi.org/10.1007/s00248-005-0046-x
Yergeau E, Sommerville DW, Maheux E, Vujanovic V, Hamel C, Whalen JK, St-Arnaud M (2006) Relationships between fusarium population structure, soil nutrient status and disease incidence in field-grown asparagus:fusarium ecology in asparagus fields. FEMS Microbiol Ecol 58:394–403. https://doi.org/10.1111/j.1574-6941.2006.00161.x
Abdellatif L, Fernandez MR, Bouzid S, Vujanovic V (2010) Characterization of virulence and PCR-DGGE profiles of Fusarium avenaceum from Western Canadian prairie ecozone of Saskatchewan. Can J Plant Pathol 32:468–480. https://doi.org/10.1080/07060661.2010.510643
Lievens B, Rep M, Thomma BP (2008) Recent developments in the molecular discrimination of formae speciales of Fusarium oxysporum. Pest Manag Sci 64:781–788. https://doi.org/10.1002/ps.1564
Clay K (1988) Fungal endophytes of grasses: a defensive mutualism between plants and fungi. Ecology 69:10–16. https://doi.org/10.2307/1943155
Jumpponen ARI, Trappe JM (1998) Dark septate endophytes: a review of facultative biotrophic root-colonizing fungi. New Phytol 140:295–310. https://doi.org/10.1046/j.1469-8137.1998.00265.x
Xia Y, DeBolt S, Dreyer J, Scott D, Williams MA (2015) Characterization of culturable bacterial endophytes and their capacity to promote plant growth from plants grown using organic or conventional practices. Front Plant Sci 6:490. https://doi.org/10.3389/fpls.2015.00490
Hermosa MR, Grondona I, Iturriaga EA, Diaz-Minguez JM, Castro C, Monte E, Garcia-Acha I (2000) Molecular characterization and identification of biocontrol isolates of Trichoderma spp. Appl Environ Microbiol 66:1890–1898. https://doi.org/10.1128/AEM.66.5.1890-1898.2000
Hirpara DG, Gajera HP, Hirapara JG, Golakiya BA (2017) Inhibition coefficient and molecular diversity of multi stress tolerant Trichoderma as potential biocontrol agent against Sclerotium rolfsii Sacc. Infect Genet Evol 55:75–92. https://doi.org/10.1016/j.meegid.2017.08.029
Jalali F, Zafari D, Salari H (2017) Volatile organic compounds of some Trichoderma spp. increase growth and induce salt tolerance in Arabidopsis thaliana. Fungal Ecol 29:67–75. https://doi.org/10.1016/j.funeco.2017.06.007
Yang J, Tian B, Liang L, Zhang K-Q (2007) Extracellular enzymes and the pathogenesis of nematophagous fungi. Appl Microbiol Biotechnol 75:21–31. https://doi.org/10.1007/s00253-007-0881-4
Vega FE, Posada F, Catherine Aime M et al (2008) Entomopathogenic fungal endophytes. Biol Control 46:72–82. https://doi.org/10.1016/j.biocontrol.2008.01.008
Jumpponen A (2001) Dark septate endophytes - are they mycorrhizal? Mycorrhiza 11:207–211. https://doi.org/10.1007/s005720100112
Zhang Q, Gong M, Yuan J et al (2017) Dark septate endophyte improves drought tolerance in sorghum. Int J Agric Biol 19:53–60. https://doi.org/10.17957/IJAB/15.0241
Thormann MN, Currah RS, Bayley SE (2004) Patterns of distribution of microfungi in decomposing bog and fen plants. Can J Bot 82:710–720
Gudiño Gomezjurado ME, de Abreu LM, Marra LM et al (2015) Phosphate solubilization by several genera of saprophytic fungi and its influence on corn and cowpea growth. J Plant Nutr 38:675–686. https://doi.org/10.1080/01904167.2014.934480
Peng X, Qiu M (2018) Meroterpenoids from ganoderma species: a review of last five years. Nat Prod Bioprospect 8:137–149. https://doi.org/10.1007/s13659-018-0164-z
Hood IA, McDougal RL, Somchit C et al (2018) Fungi decaying the wood of fallen beech (Nothofagus) trees in the South Island of New Zealand. Can J For Res 49:1–17. https://doi.org/10.1139/cjfr-2018-0179
Acknowledgments
We are grateful to Mario Laterrière and David Gagné for their assistance with the bio-informatic analysis and thank the technical staff of the AAFC research centers in Lacombe, Lethbridge and Scott for carrying out the sampling and providing useful advice.
Funding
This study received funding from the following sources: the Agricultural Bioproducts Innovation Program (ABIP), the Prairie Canola Agronomic Research Program (PCARP), the Agriculture and Agri-Food Canada Canada Canola Cluster Growing Forward Initiative, the Alberta Canola Producers Commission, the Saskatchewan Canola Development Commission, the Manitoba Canola Growers Association, the Canola Council of Canada, the Western Grains Research Foundation, and the Natural Sciences and Engineering Research Council of Canada.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Figure S1
Cumulative precipitation proportion at each site in 2013 and 2016. The fungal communities in the canola rhizosphere may have been influenced by moisture availability in July 2016. (PNG 55 kb)
Figure S2
Rarefaction curves for each rhizosphere soil sample, showing the relationship between the number of ZOTUs and the abundance of ITS sequences reads, in the 2013 dataset. The vertical black line indicates the plateau threshold for all curves. (PNG 152 kb)
Figure S3
Rarefaction curves for each rhizosphere soil sample, showing the relationship between the number of ZOTUs and the abundance of ITS sequences reads, in the 2016 dataset. The vertical black line indicates the plateau threshold for all curves. (PNG 166 kb)
ESM 1
(DOCX 53 kb)
Rights and permissions
About this article
Cite this article
Floc’h, JB., Hamel, C., Harker, K.N. et al. Fungal Communities of the Canola Rhizosphere: Keystone Species and Substantial Between-Year Variation of the Rhizosphere Microbiome. Microb Ecol 80, 762–777 (2020). https://doi.org/10.1007/s00248-019-01475-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-019-01475-8