Abstract
Molecular surveys of eukaryotic microbial communities employing high-throughput sequencing (HTS) techniques are rapidly supplanting traditional morphological approaches due to their larger data output and reduced bench work time. Here, we directly compare morphological and Illumina data obtained from the same samples, in an effort to characterize ciliate faunas from sediments in freshwater environments. We show how in silico processing affects the final outcome of our HTS analysis, providing evidence that quality filtering protocols strongly impact the number of predicted taxa, but not downstream conclusions such as biogeography patterns. We determine the abundance distribution of ciliates, showing that a small fraction of abundant taxa dominates read counts. At the same time, we advance reasons to believe that biases affecting HTS abundances may be significant enough to blur part of the underlying biological picture. We confirmed that the HTS approach detects many more taxa than morphological inspections, and highlight how the difference varies among taxonomic groups. Finally, we hypothesize that the two datasets actually correspond to different conceptions of “diversity,” and consequently that neither is entirely superior to the other when investigating environmental protists.





Similar content being viewed by others
References
Caron DA, Worden AZ, Countway PD, Demir E, Heidelberg KB (2009) Protists are microbes too: a perspective. ISME J 3:4–12
Weisse T (2008) Distribution and diversity of aquatic protists: an evolutionary and ecological perspective. Biodivers Conserv 17:243–259
Sherr EB, Sherr BF (2002) Significance of predation by protists in aquatic microbial food webs. Antonie Van Leeuwenhoek 81:293–308
Weisse T, Müller H (1998) Planktonic protozoa and the microbial food web in Lake Constance. Arch Hydrobiol 53:223–254
de Vargas C, Audic S, Henry N, et al. (2015) Eukaryotic plankton diversity in the sunlit ocean. Science 348:1261605. doi:10.1126/science.1261605
Martin C (2015) Biology’s dark matter. Curr Biol 25:R301–R307
Dolan JR, Landry MR, Ritchie ME (2013) The species-rich assemblages of tintinnids (marine planktonic protists) are structured by mouth size. ISME J 7:1237–1243
Finlay BJ, Esteban GF (1998) Freshwater protozoa: biodiversity and ecological function. Biodivers Conserv 7:1163–1186
Graham JM, Kent AD, Lauster GH, Yannarell AC, Graham LE, Triplett EW (2004) Seasonal dynamics of phytoplankton and planktonic protozoan communities in a northern temperate humic lake: diversity in a dinoflagellate dominated system. Microb Ecol 48:528–540
Bass D, Richards TA, Matthai L, Marsh V, Cavalier-Smith T (2007) DNA evidence for global dispersal and probable endemicity of protozoa. BMC Evol Biol 7:162. doi:10.1186/1471-2148-7-162
Edgcomb VP, Kysela DT, Teske A, de Vera GA, Sogin ML (2002) Benthic eukaryotic diversity in the Guaymas Basin hydrothermal vent environment. Proc Natl Acad Sci U S A 99:7658–7662
Moon-van der Staay SY, De Wachter R, Vaulot D (2001) Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409:607–610
Stoeck T, Hayward B, Taylor GT, Varela R, Epstein SS (2006) A multiple PCR-primer approach to access the microeukaryotic diversity in environmental samples. Protist 157:31–43
Behnke A, Engel M, Christen R, Nebel M, Klein RR, Stoeck T (2010) Depicting more accurate pictures of protistan community complexity using pyrosequencing of hypervariable SSU rRNA gene regions. Environ Microbiol 13:140–149
Grossmann L, Jensen M, Heider D, et al. (2016) Protistan community analysis: key findings of a large-scale molecular sampling. ISME J 10:2269–2279
Logares R, Audic S, Bass D, et al. (2014) Patterns of rare and abundant marine microbial eukaryotes. Curr Biol 24:813–821
Stoeck T, Bass D, Nebel M, Christen R, Jones MDM, Breiner HW, Richards TA (2010) Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol 1:21–31
Sunagawa S, Coelho LP, Chaffron S, et al. (2015) Structure and function of the global ocean microbiome. Science 348:1261359. doi:10.1126/science.1261359
Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ (2006) Microbial diversity in the deep sea and the underexplored “rare biodiversity”. Proc Natl Acad Sci U S A 103:12115–12120
Delmont TO, Robe P, Cecillon S, Clark IM, Constancias F, Simonet P, Hirsch PR, Vogel TM (2011) Accessing the soil metagenome for studies of microbial diversity. Appl Environ Microbiol 77:1315–1324
Fonseca VG, Nichols B, Lallias D, Quince C, Carvalho GR, Power DM, Creer S (2012) Sample richness and genetic diversity as drivers of chimera formation in nSSU metagenomic analyses. Nucleic Acids Res 40:e66. doi:10.1093/nar/gks002
Schloss PD, Gevers D, Westcott SL (2011) Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6:e27310. doi:10.1371/journal.pone.0027310
Schmidt TSB, Matias Rodrigues JF, von Mering C (2015) Limits to robustness and reproducibility in the demarcation of operational taxonomic units. Environ Microbiol 17:1689–1706
Massana R, Gobet A, Audic S, et al. (2015) Marine protist diversity in European coastal waters and sediments as revealed by high-throughput sequencing. Environ Microbiol 17:4035–4049
Stoeck T, Breiner HW, Filker S, Ostermaier V, Kammerlander B, Sonntag B (2014) A morphogenetic survey on ciliate plankton from a mountain lake pinpoints the necessity of lineage-specific barcode markers in microbial ecology. Environ Microbiol 16:430–444
Majaneva M, Hyytiäinen K, Varvio SL, Nagai S, Blomster J (2015) Bioinformatic amplicon read processing strategies strongly affect eukaryotic diversity and the taxonomic composition of communities. PLoS One 10:e0130035. doi:10.1371/journal.pone.0130035
Bachy C, Dolan JR, López-García P, Deschamps P, Moreira D (2013) Accuracy of protist diversity assessments: morphology compared with cloning and direct pyrosequencing of 18S rRNA genes and ITS regions using the conspicuous tintinnid ciliates as a case study. ISME J 7:244–255
Dunthorn M, Stoeck T, Clamp J, Warren A, Mahé F (2014) Ciliates and the rare biosphere: a review. J Eukaryot Microbiol 61:404–409
Medinger R, Nolte V, Pandey RV (2010) Diversity in a hidden world: potential and limitation of next-generation sequencing for surveys of molecular diversity of eukaryotic microorganisms. Mol Ecol 19:32–40
Pawlowski J, Esling P, Lejzerowicz F, Cedhagen T, Wilding TA (2014) Environmental monitoring through protist next-generation sequencing metabarcoding: assessing the impact of fish farming on benthic foraminifera communities. Mol Ecol Resour 14:1129–1140
Rossi A, Boscaro V, Carducci D, Serra V, Modeo L, Verni F, Fokin SI, Petroni G (2016) Ciliate communities and hidden biodiversity in freshwater biotopes of the Pistoia province (Tuscany, Italy). Eur J Protistol 53:11–19
Dias RJP, Wieloch AH, D’Agosto M (2008) The influence of environmental characteristics on the distribution of ciliates (protozoa, Ciliophora) in an urban stream of Southeast Brazil. Braz J Biol 68:287–295
Madoni P, Braghiroli S (2007) Changes in the ciliate assemblage along a fluvial system related to physical, chemical and geomorphological characteristics. Eur J Protistol 43:67–75
Lara E, Berney C, Harms H, Chatzinotas A (2007) Cultivation-independent analysis reveals a shift in ciliate 18S rRNA gene diversity in a polycyclic aromatic hydrocarbon-polluted soil. FEMS Microbiol Ecol 62:365–373
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596
Ludwig W, Strunk O, Westram R, et al. (2004) ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371
Dunthorn M, Klier J, Bunge J, Stoeck T (2012) Comparing the hyper-variable V4 and V9 regions of the small subunit rDNA for assessment of ciliate environmental diversity. J Eukaryot Microbiol 59:185–187
Dunthorn M, Otto J, Berger SA et al. (2014) Placing environmental next-generation sequencing amplicons from microbial eukaryotes into a phylogenetic context. Mol Biol Evol 31:993–1009
Caporaso JG, Kuczynski J, Stombaugh J, et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998
Lynn DH (2008) The ciliated protozoa. Characterization, classification, and guide to the literature, 3rd edn. Springer, Dordrecht
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron 4:9
Richards TA, Leonard G, Mahé F, et al. (2015) Molecular diversity and distribution of marine fungi across 130 European environmental samples. Proc Biol Sci 282:20152243. doi:10.1098/rspb.2015.2243
Esling P, Lejzerowicz F, Pawlowski J (2015) Accurate multiplexing and filtering for high-throughput amplicon-sequencing. Nucleic Acids Res 43:2513–2524
Filker S, Sommaruga R, Vila I, Stoeck T (2016) Microbial eukaryote plankton communities of high-mountain lakes from three continents exhibit strong biogeographic patterns. Mol Ecol 25:2286–2301
Massana R, del Campo J, Sieracki ME, Audic S, Logares R (2014) Exploring the uncultured microeukaryote majority in the oceans: reevaluation of ribogroups within stramenopiles. ISME J 8:854–866
del Campo J, Mallo D, Massana R, de Vargas C, Richards TA, Ruiz-Trillo I (2015) Diversity and distribution of unicellular opisthokonts along the European coast analysed using high-throughput sequencing. Environ Microbiol 17:3195–3207
Egge ES, Johannessen TV, Andersen T, Eikrem W, Bittner L, Larsen A, Sandaa RA, Edvardsen B (2015) Seasonal diversity and dynamics of haptophytes in the Skagerrak, Norway, explored by high-throughput sequencing. Mol Ecol 24:3026–3042
Nanjappa D, Audic S, Romac S, Kooistra WHCF, Zingone A (2014) Assessment of species diversity and distribution of an ancient diatom lineage using a DNA metabarcoding approach. PLoS One 9:e103810. doi:10.1371/journal.pone.0103810
Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120
Chao A, Li PC, Agatha S, Foissner W (2006) A statistical approach to estimate soil ciliate diversity and distribution based on data from five continents. Oikos 114:479–493
Nebel M, Pfabel C, Stock A, Dunthorn M, Stoeck T (2011) Delimiting operational taxonomic units for assessing ciliate environmental diversity using small-subunit rRNA gene sequences. Environ Microbiol Rep 3:154–158
Nielsen KM, Johnsen PJ, Bensasson D, Daffonchio D (2007) Release and persistence of extracellular DNA in the environment. Environ Biosaf Res 6:37–53
Vlassov VV, Laktionov PP, Rykova EY (2007) Extracellular nucleic acids. BioEssays 29:654–667
Lynch MDJ, Neufeld JD (2015) Ecology and exploration of the rare biosphere. Nat Rev Microbiol 13:217–229
Pedrós-Alió C (2006) Marine microbial diversity: can it be determined? Trends Microbiol 14:257–263
Zhu F, Massana R, Not F, Marie D, Vaulot D (2005) Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol Ecol 52:79–92
Gong J, Dong J, Liu X, Massana R (2013) Extremely high copy numbers and polymorphisms of the rDNA operon estimated from single cell analysis of oligotrich and peritrich ciliates. Protist 164:369–379
Prescott DM (1994) The DNA of ciliated protozoa. Microbiol Rev 58:233–267
Riley JL, Katz LA (2001) Widespread distribution of extensive chromosomal fragmentation in ciliates. Mol Biol Evol 18:1372–1377
Visco JA, Apothéloz-Perret-Gentil L, Cordonier A, Esling P, Pillet L, Pawlowski J (2015) Environmental monitoring: inferring the Diatom Index from next-generation sequencing data. Environ Sci Technol 49:7597–7605
Fenchel T (1987) Ecology of protozoa. The biology of free-living phagotrophic protists. Springer-Verlag, Berlin
Acknowledgements
The authors wish to thank Simone Gabrielli for his help in graphical artwork, Valentina Serra and Daniela Carducci for assistance during sampling, and Brad Jones for language editing.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
.
Online Resource 1
Additional experimental procedures details. (PDF 90 kb)
.
Online Resource 2
Number of morphological and Illumina OTUs, divided by ciliate class, in each of the 12 samples. Contig abundances are also reported. (PDF 2444 kb)
.
Online Resource 3
Additional pipeline comparisons and statistical test results. (PDF 2468 kb)
.
Online Resource 4
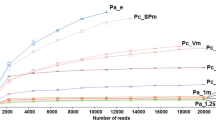
Rarefaction curves calculated on the 12 samples. (TIFF 356 kb)
.
Online Resource 5
Number of contigs (A) and OTUs (B) assigned to each ciliate subclass in the entire analysis (pipeline U.s.). Some ciliate classes are not subdivided in subclasses [41]; in those cases, we maintained class names (underlined). (TIFF 1167 kb)
.

Online Resource 6
Relative number of OTUs per sample, divided by class, according to the morphological analysis (A) and the HTS survey (pipeline U.s.) (B). (JPEG 224 kb)
.
Online Resource 7
nMDS analysis performed on presence/absence morphological data from all samples, employing Bray-Curtis distances. Symbols represent the 4 sampling sites according to the legend. The gradient analysis was performed on abiotic factors, represented by vectors. A similar graph including more samples from the same and other sites belonging to the investigated area can be consulted in Rossi et al. [31]. T, temperature; O, oxygen concentration; A, altitude. (EPS 311 kb)
Rights and permissions
About this article
Cite this article
Boscaro, V., Rossi, A., Vannini, C. et al. Strengths and Biases of High-Throughput Sequencing Data in the Characterization of Freshwater Ciliate Microbiomes. Microb Ecol 73, 865–875 (2017). https://doi.org/10.1007/s00248-016-0912-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-016-0912-8