Abstract
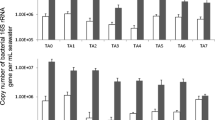
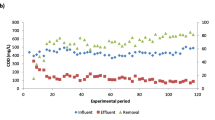
In this study, changes in eukaryotic community structure and water quality were investigated in an aerobic trickling filter (down-flow hanging sponge, DHS) treating domestic sewage under different organic loading rates (OLRs). The OLR clearly influenced both sponge pore water quality and relative flagellates and ciliates (free-swimming, carnivorous, crawling, and stalked protozoa) abundances in the retained sludge. Immediately after the OLR was increased from 1.05 to 1.97 kg chemical oxygen demand (COD) m−3 day−1, COD and NH4 +-N treatment efficiencies both deteriorated, and relative flagellates and ciliates abundances then increased from 2–8 % to 51–65 % total cells in the middle-bottom part of the DHS reactor. In a continuous operation at a stable OLR (2.01 kg COD m−3 day−1), effluent water quality improved, and relative flagellates and ciliates abundances decreased to 15–46 % total cells in the middle-bottom part of the DHS reactor. This result may indicate that flagellates and ciliates preferentially graze on dispersed bacteria, thus, stabilizing effluent water quality. Additionally, to investigate eukaryotic community structure, clone libraries based on the 18S ribosomal ribonucleic acid (rRNA) gene of the retained sludge were constructed. The predominant group was Nucletmycea phylotypes, representing approximately 29–56 % total clones. Furthermore, a large proportion of the clones had <97 % sequence identity in the NCBI database. This result indicates that phylogenetically unknown eukaryotes were present in the DHS reactor. These results provide insights into eukaryotic community shift in the DHS reactor treating domestic sewage.






Similar content being viewed by others
References
Canals O, Salvadó H, Auset M, Hernández C, Malfeito JJ (2013) Microfauna communities as performance indicators for A/O shortcut biological nitrogen removal moving-bed biofilm reactor. Water Res 47:3141–3150
Curds CR, Fey GJ (1969) The effect of ciliated protozoa on the fate of Escherichia coli in the activated-sludge process. Water Res 3:853–867
Lapinski J, Tunnacliffe A (2003) Reduction of suspended biomass in municipal wastewater using bdelloid rotifers. Water Res 37:2027–2034
Ravva SV, Sarreal CZ, Mandrell RE (2010) Identification of protozoa in dairy lagoon wastewater that consume Escherichia coli O157: H7 preferentially. PLoS ONE 5(12):e15671
Moreno AM, Matz C, Kjelleberg S, Manefield M (2010) Identification of ciliate grazers of autotrophic bacteria in ammonia-oxidizing activated sludge by RNA isotope probing. Appl Environ Microbiol 76(7):2203–2211
Evans TN, Seviour RJ (2012) Estimating biodiversity of fungi in activated sludge communities using culture-independent methods. Microbiol Ecol 63:773–786
Matsunaga K, Kubota K, Harada H (2014) Molecular diversity of eukaryotes in municipal wastewater treatment processes as revealed by 18S rRNA gene analysis. Microbes Environ 29(4):401–407
Hayatsu M, Tago K, Saito M (2008) Various players in the nitrogen cycle: diversity and functions of the microorganisms involved in nitrification and denitrification. Soil Sci Plant Nutr 54:33–45
Baldrian P, Valášková V (2008) Degradation of cellulose by basidiomycetous fungi. FEMS Microbiol Rev 32:501–521
Drake JF, Tsuchiya HM (1977) Growth kinetics of Colpoda steinii on Escherichia coli. Appl Environ Microbiol 34(1):18–22
Taylor WD (1978) Growth responses of ciliate protozoa to the abundance of their bacterial prey. Microb Ecol 4:207–214
Marsh TL, Liu WT, Forney LJ, Cheng H (1988) Beginning a molecular analysis of the eukaryal community in activated sludge. Water Sci Technol 37(4–5):455–460
Shimano S, Sanbe M, Kasahara Y (2008) Linkage between light microscopic observations and molecular analysis by single-cell PCR for ciliates. Microbes Environ 23(4):356–359
Shimano S, Sambe M, Kasahara Y (2012) Application of nested PCR-DGGE (denaturing gradient gel electrophoresis) for the analysis of ciliate communities in soils. Microbes Environ 27(2):136–141
Zhu F, Massana R, Not F, Marie D, Vaulot D (2005) Mapping of picoeukaryotes in marine ecosystem with quantitative PCR of the 18S rRNA gene. FEMS Microbiol Ecol 52:79–92
Machdar I, Harada H, Ohashi A, Sekiguchi Y, Okui H, Ueki K (1997) A novel and cost-effective sewage treatment system consisting of UASB pre-treatment and aerobic post-treatment units for developing countries. Water Sci Technol 36(12):189–197
Uemura S, Harada H (2010) Application of UASB technology for sewage treatment with a novel post-treatment process. In: Fang HHP (ed) Environmental anaerobic technology. Imperial College Press, London, pp 91–112
Agrawal LK, Ohashi Y, Mochida E, Okui H, Ueki Y, Harada H, Ohashi A (1997) Treatment of raw sewage in a temperate climate using a UASB reactor and the hanging sponge cubes process. Water Sci Technol 36(6–7):433–440
Yoochatchaval W, Onodera T, Sumino H, Yamaguchi T, Mizuochi M, Okadera T, Syutsubo K (2014) Development of a down-flow hanging sponge reactor for the treatment of low-strength sewage. Water Sci Technol 70(4):656–663
Tawfik A, El-Gohary F, Ohashi A, Harada H (2006) The influence of physical-chemical and biological factors on the removal of faecal coliform through down-flow hanging sponge (DHS) system treating UASB reactor effluent. Water Res 40:1877–1883
Kubota K, Hayashi M, Matsunaga K, Iguchi A, Ohashi A, Li YY, Yamaguchi Y, Harada H (2014) Microbial community composition of a down-flow hanging sponge (DHS) reactor combined with an up-flow anaerobic sludge blanket (UASB) reactor for the treatment for the treatment of municipal sewage. Bioresour Technol 151:144–150
Onodera T, Matsunaga K, Kubota K, Taniguchi R, Harada H, Syutsubo K, Okubo T, Uemura S, Araki N, Yamada M, Yamauchi M, Yamaguchi T (2013) Characterization of the retained sludge in a down-flow hanging sponge (DHS) reactor with emphasis on its low excess sludge production. Bioresour Technol 136:169–175
Hatamoto M, Ohtsuki K, Maharjan N, Ono S, Dehama K, Sakamoto K, Takahashi M, Yamaguchi T (2016) Performance evaluation of the sulfur-redox-reaction-activated up-flow anaerobic sludge blanket and down-flow hanging sponge anaerobic/anoxic sequencing batch reactor system for municipal sewage treatment. Bioresour Technol 204:171–176
Lim EL, Amaral LA, Caron DA, DeLong EF (1993) Application of rRNA-based probes for observing marine nanoplanktonic protists. Appl Environ Microbiol 59(5):1647–1655
Medlin L, Elwood HJ, Stickel S, Sogin ML (1988) The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71:491–499
Karnati SKR, Yu Z, Sylvester JT, Dehority BA, Morrison M, Firkins JL (2003) Technical note: specific PCR amplification of protozoal 18S rDNA sequence from DNA extracted from ruminal samples of cows. J Anim Sci 81:812–815
López-García P, Rodriguez-Valera F, Pedrós-Alió C, Moreira D (2001) Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 409:603–607
Pruesse E, Quast C, Knittel K, Fuch BM, Ludwig W, Peplies J, Glöckner O (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35(21):7188–7196
Good IJ (1953) The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264
Lara E, Moreira D, López-García P (2010) The environmental clade LKM11 and Rozella form the deepest branching clade of fungi. Protist 161:116–121
Brown MW, Spiegel FW, Silberman JD (2009) Phylogeny of the “forgotten” cellular slime mold, Fonticula alba, reveals a key evolutionary branch within Opisthokonta. Mol Biol Evol 26(12):2699–2709
Weber SD, Hofmann A, Pilhofer M, Wanner G, Agerer R, Ludwig W, Schleifer KH, Fried J (2009) The diversity of fungi in aerobic sewage granules assessed by 18S rRNA gene and ITS sequence analyses. FEMS Microbiol Ecol 68:246–254
Syutsubo K, Yoochatchaval W, Tsushima I, Araki N, Kubota K, Onodera T, Takahashi M, Yamaguchi T, Yoneyama Y (2011) Evaluation of sludge properties in a pilot-scale UASB reactor for sewage treatment in a temperature region. Water Sci Technol 64(10):1959–1966
Madoni P (1994) Estimates of ciliated protozoa biomass in activated sludge and biofilm. Bioresour Technol 48:245–249
Matz C, Kjelleberg S (2005) Off the hook—how bacteria survive protozoan grazing. Trends Microbiol 13(7):302–307
Pajdak-Stós A, Fiałkowska E, Fyda J, Babko R (2010) Resistance of nitrifiers inhabiting activated sludge to ciliate grazing. Water Sci Technol 61(3):573–580
Machdar I, Sekiguchi Y, Sumino H, Ohashi A, Harada H (2000) Combination of a UASB reactor and a curtain type DHS (downflow hanging sponge) reactor as a cost-effective sewage treatment system for developing countries. Water Sci Technol 42(3–4):83–88
Dubber D, Gray NF (2011) The effect of anoxia and anaerobia on ciliate community in biological nutrient removal system using laboratory-scale sequencing batch reactor. Water Res 45:2213–2226
Snaidr J, Amann R, Huber I, Ludwig W, Schleifer KH (1997) Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol 63(7):2884–2896
Salvadó H, Grancia MP (1993) Determination of organic loading rate of activated sludge plants based on protozoan analysis. Water Res 27(5):891–895
Liu J, Yang M, Qi R, An W, Zhou J (2008) Comparative study of protozoan communities in full-scale MWTPs in Beijing related to treatment processes. Water Res 42:1907–1918
Madoni P, Davoli D, Chierici E (1993) Comparative analysis of the activated sludge microfauna in several sewage treatment works. Water Res 27(9):1485–1491
Zhou K, Xu M, Dai J, Cao H (2006) The microfauna communities and operational monitoring of an activated sludge plant in China. Eur J Protistol 42:291–295
Suarez C, Persson F, Hermansson M (2015) Predation of nitritation-anammox biofilms used for nitrogen removal from wastewater. FEMS Microbiol Ecol 91(11):1–9
Okubo T, Onodera T, Uemura S, Yamaguchi T, Ohashi A, Harada H (2015) On-site evaluation of the performance of a full-scale down-flow hanging sponge reactor as a post-treatment process of an up-flow anaerobic sludge blanket reactor for treating sewage in India. Bioresour Technol 194:156–164
Tyagi VK, Subramaniyan S, Kazmi AA, Chopra AK (2008) Microbial community in conventional and extended aeration activated sludge plants in India. Ecol Indic 8(5):550–554
Tsuruta S, Takaya N, Zhang L, Shoun H, Kimura K, Hamamoto M, Nakase T (1998) Denitrification by yeast and occurrence of cytochrome P450nor in Trichosporon cutanum. FEMS Microbiol Lett 168:105–110
Acknowledgments
The authors thank the staff at the municipal sewage works at Tsuchiura city, Ibaraki prefecture, Japan. This research is supported by NIES Issue-Oriented Research Program, the Japan Society for the Promotion of Science (KAKENHI Grant Number 15J10199), and Environment Research and Technology Development Fund (1-1603) of the Ministry of the Environment, Japan.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 4532 kb)
Rights and permissions
About this article
Cite this article
Miyaoka, Y., Hatamoto, M., Yamaguchi, T. et al. Eukaryotic Community Shift in Response to Organic Loading Rate of an Aerobic Trickling Filter (Down-Flow Hanging Sponge Reactor) Treating Domestic Sewage. Microb Ecol 73, 801–814 (2017). https://doi.org/10.1007/s00248-016-0871-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-016-0871-0