Abstract
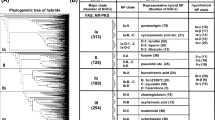
The chemical ecology and biotechnological potential of metabolites from endophytic and rhizosphere fungi are receiving much attention. A collection of 17 sugarcane-derived fungi were identified and assessed by PCR for the presence of polyketide synthase (PKS) genes. The fungi were all various genera of ascomycetes, the genomes of which encoded 36 putative PKS sequences, 26 shared sequence homology with β-ketoacyl synthase domains, while 10 sequences showed homology to known fungal C-methyltransferase domains. A neighbour–joining phylogenetic analysis of the translated sequences could group the domains into previously established chemistry-based clades that represented non-reducing, partially reducing and highly reducing fungal PKSs. We observed that, in many cases, the membership of each clade also reflected the taxonomy of the fungal isolates. The functional assignment of the domains was further confirmed by in silico secondary and tertiary protein structure predictions. This genome mining study reveals, for the first time, the genetic potential of specific taxonomic groups of sugarcane-derived fungi to produce specific types of polyketides. Future work will focus on isolating these compounds with a view to understanding their chemical ecology and likely biotechnological potential.





Similar content being viewed by others
References
Amnuaykanjanasin A, Phonghanpot S, Sengpanich N, Cheevadhanarak S, Tanticharoen M (2009) Insect-specific polyketide synthases (PKSs), potential PKS-nonribosomal peptide synthetase hybrids, and novel PKS clades in tropical fungi. Appl Environ Microbiol 75:3721–3732
Amnuaykanjanasin A, Punya J, Paungmoung P, Rungrod A, Tachaleat A, Pongpattanakitshote S, Cheevadhanarak S, Tanticharoen M (2005) Diversity of type I polyketide synthase genes in the wood-decay fungus Xylaria sp. BCC 1067. FEMS Microbiol Lett 251:125–136
Ansari MZ, Sharma J, Gokhale RS, Mohanty D (2008) In silico analysis of methyltransferase domains involved in biosynthesis of secondary metabolites. BMC Bioinformatics 9:454
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201
Atoui A, Dao HP, Mathieu F, Lebrihi A (2006) Amplification and diversity analysis of ketosynthase domains of putative polyketide synthase genes in Aspergillus ochraceus and Aspergillus carbonarius producers of ochratoxin A. Mol Nutr Food Res 50:488–493
Bach TJ, Lichtenthaler HK (1983) Inhibition by mevinolin of plant growth, sterol formation and pigment acumulation. Physiol Plant 59:50–60
Ballance DJ (1986) Sequences important for gene expression in filamentous fungi. Yeast 2:229–236
Bills G, Overy D, Genilloud O, Pelaez F (2009) Contribution of pharmaceutical antibiotics and secondary metabolite discovery to the understanding of microbial defence and antagonism. In: White JM, Torres MS (eds) Defensive mutualism in microbial symbiosis, vol. 27. CRC, Boca Raton
Bingle LE, Simpson TJ, Lazarus CM (1999) Ketosynthase domain probes identify two subclasses of fungal polyketide synthase genes. Fungal Genet Biol 26:209–223
Cox RJ (2007) Polyketides, proteins and genes in fungi: programmed nano-machines begin to reveal their secrets. Org Biomol Chem 5:2010–2026
Da Silva M, Passarini MR, Bonugli RC, Sette LD (2008) Cnidarian-derived filamentous fungi from Brazil: isolation, characterisation and RBBR decolourisation screening. Environ Technol 29:1331–1339
Eriksson OE, Hawksworth DL (2003) Saccharicola, a new genus for two Leptosphaeria species on sugar cane. Mycologia 95:426–433
Galgoczy L, Papp T, Lukacs G, Leiter E, Pocsi I, Vagvolgyi C (2007) Interactions between statins and Penicillium chrysogenum antifungal protein (PAF) to inhibit the germination of sporangiospores of different sensitive Zygomycetes. FEMS Microbiol Lett 270:109–115
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118
Ginolhac A, Jarrin C, Gillet B, Robe P, Pujic P, Tuphile K, Bertrand H, Vogel TM, Perriere G, Simonet P, Nalin R (2004) Phylogenetic analysis of polyketide synthase I domains from soil metagenomic libraries allows selection of promising clones. Appl Environ Microbiol 70:5522–5527
Gontang EA, Gaudencio SP, Fenical W, Jensen PR (2010) Sequence-based analysis of secondary-metabolite biosynthesis in marine actinobacteria. Appl Environ Microbiol 76:2487–2499
Grube M, Blaha J (2003) On the phylogeny of some polyketide synthase genes in the lichenized genus Lecanora. Mycol Res 107:1419–1426
Guerrero A, Rosell G (2005) Biorational approaches for insect control by enzymatic inhibition. Curr Med Chem 12:461–469
Henson JM, Butler MJ, Day AW (1999) The dark sede of the mycelium: melanins of phytopathogenic fungi. Annu Rev Phytopathol 37:447–471
Hertweck C (2009) The biosynthetic logic of polyketide diversity. Angew Chem Int Ed Engl 48:4688–4716
Hoffmeister D, Keller NP (2007) Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat Prod Rep 24:393–416
Hopwood DA, Khosla C (1992) Genes for polyketide secondary metabolic pathways in microorganisms and plants. Ciba Found Symp 171:88–106, discussion 106–112
Howard RJ, Ferrari MA, Roach DH, Money NP (1991) Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc Natl Acad Sci USA 88:11281–11284
Huang CS, Chang SL (1972) Leafinfection with citrus black spot and perithecial development in relation to ascospore discharge of Guignardia citricarpa Kiely. J Taiwan Agric Res 21:256–263
Huang W, Jia J, Edwards P, Dehesh K, Schneider G, Lindqvist Y (1998) Crystal structure of beta-ketoacyl-acyl carrier protein synthase II from E. coli reveals the molecular architecture of condensing enzymes. EMBO J 17:1183–1191
Kagan RM, Clarke S (1994) Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes. Arch Biochem Biophys 310:417–427
Katz JE, Dlakic M, Clarke S (2003) Automated identification of putative methyltransferases from genomic open reading frames. Mol Cell Proteomics 2:525–540
Kelley LA, Sternberg MJ (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4:363–371
Kim TK, Hewavitharana AK, Shaw PN, Fuerst JA (2006) Discovery of a new source of rifamycin antibiotics in marine sponge actinobacteria by phylogenetic prediction. Appl Environ Microbiol 72:2118–2125
Kroken S, Glass NL, Taylor JW, Yoder OC, Turgeon BG (2003) Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc Natl Acad Sci USA 100:15670–15675
Kupfer DM, Drabenstot SD, Buchanan KL, Lai H, Zhu H, Dyer DW, Roe BA, Murphy JW (2004) Introns and splicing elements of five diverse fungi. Eukaryot Cell 3:1088–1100
Langfelder K, Streibel M, Jahn B, Haase G, Brakhage AA (2003) Biosynthesis of fungal melanins and their importance for human pathogenic fungi. Fungal Genet Biol 38:143–158
Lee T, Yun SH, Hodge KT, Humber RA, Krasnoff SB, Turgeon GB, Yoder OC, Gibson DM (2001) Polyketide synthase genes in insect- and nematode-associated fungi. Appl Microbiol Biotechnol 56:181–187
Macreadie IG, Johnson G, Schlosser T, Macreadie PI (2006) Growth inhibition of Candida species and Aspergillus fumigatus by statins. FEMS Microbiol Lett 262:9–13
Magnani GS, Didonet CM, Cruz LM, Picheth CF, Pedrosa FO, Souza EM (2010) Diversity of endophytic bacteria in Brazilian sugarcane. Genet Mol Res 9:250–258
Martin JL, McMillan FM (2002) SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr Opin Struct Biol 12:783–793
Mathieu M, Zeelen JP, Pauptit RA, Erdmann R, Kunau WH, Wierenga RK (1994) The 2.8 A crystal structure of peroxisomal 3-ketoacyl-CoA thiolase of Saccharomyces cerevisiae: a five-layered alpha beta alpha beta alpha structure constructed from two core domains of identical topology. Structure 2:797–808
Mayer KM, Ford J, Macpherson GR, Padgett D, Volkmann-Kohlmeyer B, Kohlmeyer J, Murphy C, Douglas SE, Wright JM, Wright JL (2007) Exploring the diversity of marine-derived fungal polyketide synthases. Can J Microbiol 53:291–302
Mendes R, Pizzirani-Kleiner AA, Araujo WL, Raaijmakers JM (2007) Diversity of cultivated endophytic bacteria from sugarcane: genetic and biochemical characterization of Burkholderia cepacia complex isolates. Appl Environ Microbiol 73:7259–7267
Metz JG, Roessler P, Facciotti D, Levering C, Dittrich F, Lassner M, Valentine R, Lardizabal K, Domergue F, Yamada A, Yazawa K, Knauf V, Browse J (2001) Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science 293:290–293
Miller DJ, Ouellette N, Evdokimova E, Savchenko A, Edwards A, Anderson WF (2003) Crystal complexes of a predicted S-adenosylmethionine-dependent methyltransferase reveal a typical AdoMet binding domain and a substrate recognition domain. Protein Sci 12:1432–1442
Nicholson TP, Rudd BA, Dawson M, Lazarus CM, Simpson TJ, Cox RJ (2001) Design and utility of oligonucleotide gene probes for fungal polyketide synthases. Chem Biol 8:157–178
Olsen JG, Kadziola A, von Wettstein-Knowles P, Siggaard-Andersen M, Larsen S (2001) Structures of beta-ketoacyl-acyl carrier protein synthase I complexed with fatty acids elucidate its catalytic machinery. Structure 9:233–243
Peres NA, Harakava R, Carroll GC, Adaskaveg JE, Timmer LW (2007) Comparisonof molecular procedures for detection and identification of Guignardia citricarpa and G. mangiferae. Plant Dis 91:525–531
Sauer M, Lu P, Sangari R, Kennedy S, Polishook J, Bills G, An Z (2002) Estimating polyketide metabolic potential among nonsporulatingfungal endophytes of Vaccinium macrocarpon. Mycol Res 1006:460–470
Schmitt I, Kautz S, Lumbsch HT (2008) 6-MSAS-like polyketide synthase genes occur in lichenized ascomycetes. Mycol Res 112:289–296
Schmitt I, Martin MP, Kautz S, Lumbsch HT (2005) Diversity of non-reducing polyketide synthase genes in the Pertusariales (lichenized Ascomycota): a phylogenetic perspective. Phytochemistry 66:1241–1253
Schulz B, Boyle C (2005) The endophytic continuum. Mycol Res 109:661–686
Schumann J, Hertweck C (2006) Advances in cloning, functional analysis and heterologous expression of fungal polyketide synthase genes. J Biotechnol 124:690–703
Soanes DM, Alam I, Cornell M, Wong HM, Hedeler C, Paton NW, Rattray M, Hubbard SJ, Oliver SG, Talbot NJ (2008) Comparative genome analysis of filamentous fungi reveals gene family expansions associated with fungal pathogenesis. PLoS One 3:e2300
Stierle A, Strobel G, Stierle D (1993) Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of Pacific yew. Science 260:214–216
Stuart RM, Romao AS, Pizzirani-Kleiner AA, Azevedo JL, Araujo WL (2010) Culturable endophytic filamentous fungi from leaves of transgenic imidazolinone-tolerant sugarcane and its non-transgenic isolines. Arch Microbiol 192:307–313
Tan RX, Zou WX (2001) Endophytes: a rich source of functional metabolites. Nat Prod Rep 18:448–459
Varga J, Rigo K, Kocsube S, Farkas B, Pal K (2003) Diversity of polyketide synthase gene sequences in Aspergillus species. Res Microbiol 154:593–600
Weissman KJ (2009) Introduction to polyketide biosynthesis. Methods Enzymol 459:3–16
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal DNA genes for phylogenies. In: Innis MA, Gelfand DH, Snisky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic, New York, pp 312–315
Acknowledgements
The authors wish to thank Dr. Léia Cécilia de Lima Fávaro and Dr. Aline Silva Romão from ESLQ/USP for the fungal isolates used in this study. We also thank Prof. Russell J. Cox from the University of Bristol for his advice during the early stages of this work and to Dr. Fernando Peláez and Dr. Paul Long for their critical review. This project was funded by The Academy of Science for the Developing World (TWAS), Conselho Nacional de Desenvolvimento Cientifico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP).
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Materials
Below is the link to the electronic supplementary material.
Supplementary Material 1
(PDF 118 kb)
Supplementary Material 2
(PDF 85 kb)
Supplementary Material 3
(PDF 1140 kb)
Rights and permissions
About this article
Cite this article
Rojas, J.D., Sette, L.D., de Araujo, W.L. et al. The Diversity of Polyketide Synthase Genes from Sugarcane-Derived Fungi. Microb Ecol 63, 565–577 (2012). https://doi.org/10.1007/s00248-011-9938-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-011-9938-0