Abstract
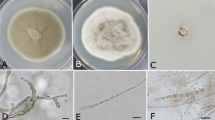
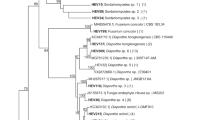
The diversity of endophytic filamentous fungi from leaves of transgenic imidazolinone-tolerant sugarcane plants and its isoline was evaluated by cultivation followed by amplified rDNA restriction analysis (ARDRA) of randomly selected strains. Transgenic and non-transgenic cultivars and their crop management (herbicide application or manual weed control) were used to assess the possible non-target effects of genetically modified sugarcane on the fungal endophytic community. A total of 14 ARDRA haplotypes were identified in the endophytic community of sugarcane. Internal transcribed spacer (ITS) sequencing revealed a rich community represented by 12 different families from the Ascomycota phylum. Some isolates had a high sequence similarity with genera that are common endophytes in tropical climates, such as Cladosporium, Epicoccum, Fusarium, Guignardia, Pestalotiopsis and Xylaria. Analysis of molecular variance indicated that fluctuations in fungal population were related to both transgenic plants and herbicide application. While herbicide applications quickly induced transient changes in the fungal community, transgenic plants induced slower changes that were maintained over time. These results represent the first draft on composition of endophytic filamentous fungi associated with sugarcane plants. They are an important step in understanding the possible effects of transgenic plants and their crop management on the fungal endophytic community.


Similar content being viewed by others
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Andreote FD, Mendes R, Dini-Andreote F, Rosseto PB, Labate CA, Pizzirani-Kleiner AA, van Elsas JD, Azevedo JL, Araújo WL (2008) Transgenic tobacco revealing altered bacterial diversity in the rhizosphere during early development. Antonie Leeuwenhoek 93:415–424
Andrews JH, Harris RF (2000) The ecology and biogeography of microorganisms on plant surfaces. Annu Rev Phytopathol 38:145–180
Arnold AE, Maynard Z, Gilbert GS, Coley PD, Kursar TA (2000) Are tropical fungal endophytes hyperdiverse? Ecol Lett 3:267–274
Azevedo JL, Araújo WL (2003) Genetically modified crops: environmental and human health concerns. Mutat Res 544:223–233
Azevedo JL, Araújo WL (2007) Diversity and applications of endophytic fungi isolated from tropical plants. In: Ganguli BN, Deshmukh SK (eds) Fungi: multifaceted microbes. Anamaya Publishers, New Delhi, India & CRC Press, Boca Raton, USA, pp 189–207
Baayen RP, Bonants PJM, Verkley G, Carroll GC, van der Aa HA, de Weerdt M et al (2002) Nonpathogenic isolates of the citrus black spot fungus, Guignardia citricarpa, identified as a cosmopolitan endophyte of wood plants, G. mangiferae (Phyllosticta capitalensis). Phytopathol 92:464–477
Canadian Food Inspection Agency [CFIA] (2003) Decision document DD2003–44 determination of the safety of BASF’s imazamox tolerant (ClearfieldTM) wheat AP602CL. Nepean, Ontario
Canadian Food Inspection Agency–CFIA (2004) Decision document DD2004–47 determination of the safety of BASF’s imazamox tolerant (ClearfieldTM) wheat AP205CL. Nepean, Ontario
Castaldini M, Turrini A, Sbrana C, Benedetti A, Marchionni M, Mocali S et al (2005) Impact of Bt corn on rhizospheric and soil eubacterial communities and on beneficial mycorrhizal symbiosis in experimental microcosms. Appl Environ Microbiol 71:6719–6729
Companhia Nacional de Abastecimento–CONAB (2009) 2cana_de_acucar.pdf. In: Acompanhamento da Safra Brasileira. Avaiable via. http://www.conab.gov.br/conabweb/download/safra/2cana_de_acucar.pdf. Accessed 10 Oct 2009
Dunfield KE, Germida JJ (2001) Diversity of bacterial communities in the rhizosphere and root interior of field-grown genetically modified Brassica napus. FEMS Microbiol Ecol 38:1–9
Dunfield KE, Germida JJ (2004) Impact of genetically modified crops on soil- and plant-associated microbial communities. J Environ Qual 33:806–815
Excoffier L, Smouse PE, Quattro M (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Götz M, Nirenberg H, Krause S, Draeger HS, Buchner A, Lottmann J, Berg G, Smalla K (2006) Fungal endophytes in potato roots studied by traditional isolation and cultivation-independent DNA-based methods. FEMS Microbiol Ecol 58:404–413
Hails RS (2000) Genetically modified plants—the debate continues. Trends Ecol Evol 15:14–18
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian protein metabolism. Academic Press, New York, USA, pp 21–132
Kremer RJ, Donald PA, Keaster AJ, Minor HC (2000) Herbicide impact on Fusarium spp. and soybean cyst nematode in glyphosate-tolerant soybean. Annual meetings abstracts. ASA, CSSA and SSSA, Madison, WI, p 257
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Kuwalchuk GA, Bruinsma M, Van Veen JA (2003) Assessing responses of soil microorganisms to GM plants. Trends Ecol Evol 18:403–410
Liu B, Zeng Q, Yan F, Xu H, Xu C (2005) Effects of transgenic plants on soil microorganisms. Plant Soil 271:1–13
Mendes R, Pizzirani-Kleiner AA, Araújo WL, Raaijmakers JM (2007) Diversity of cultivated endophytic bacteria from sugarcane: genetic and biochemical characterization of Burkholderia cepacia complex isolates. Appl Environ Microbiol 73:7259–7267
Pan SS, Duggleby RG (1999) Expression, purification, characterization, and reconstitution of the large and small subunits of yeast acetohydroxyacid synthase. Biochemistry 38:5222–5231
Peterson RKD, Shama LM (2005) A comparative risk assessment of genetically engineered, mutagenic, and conventional wheat production systems. Transgenic Res 14:859–875
Raeder U, Broda P (1985) Rapid preparation of DNA from filamentous fungi. Lett Appl Microbiol 1:17–20
Santamaría J, Bayman P (2005) Fungal epiphytes and endophytes of coffee leaves (Coffea arabica). Microb Ecol 50:1–8
Schulz B, Boyle C (2005) The endophytic continuum. Mycol Res 109:661–686
Seaby DA, Schaible RC (2000) Preventing Heterobasidion annosum from colonizing chemically thinned Sitka spruce steams. Forestry 73:351–359
Selosse MA, Baudoin E, Vandenkoornhuyse P (2004) Symbiotic microorganisms, a key for ecological success and protection of plants. C R Biologies 327:639–648
Siciliano SD, Germida JJ (1999) Taxonomic diversity of bacteria associated with the roots of field-grown transgenic Brassica napus cv. Quest compared to the non-transgenic B. napus cv. Excel and B. rapa cv. Parkland. FEMS Microbiol Ecol 29:263–272
Suryanarayanan TS, Venkatesan G, Murali TS (2003) Endophytic fungal communities in leaves of tropical forest trees: diversity and distribution patterns. Curr Sci 85:489–493
Thioulouse J, Chessel D, Dolédec S, Olivier JM (1997) ADE-4: a multivariate analysis and graphical display software. Stat Comput 7:75–83
Turrini A, Sbrana C, Nuti MP, Pietrangeli BM, Giovannetti M (2004) Development of a model system to assess the impact of genetically modified corn and aubergine plants on arbuscular mycorrhizal fungi. Plant Soil 266:69–75
Wyss GS, Charudattan R, Rosskopf EN, Littell RC (2004) Effects of selected pesticides and adjuvants on germination and vegetative growth of Phomopsis amaranthicola, a biocontrol agent for Amaranthus spp. Weed Res 44:469–482
Zucchi MI, Arizono H, Morais VA, Fungaro MHP, Vieira MLC (2002) Genetic instability of sugarcane plants derived from meristem cultures. Genet Mol Biol 25:91–96
Acknowledgments
We thank Dr. Eugênio César Ulian (Centro de Tecnologia Canavieira) for transgenic and non-transgenic sugarcane plants and crop sites. This work was supported by FAPESP (Proc. no. 02/14143-3). We also thank CAPES for providing the fellowship to R.M.S.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Erko Stackebrandt.
Rights and permissions
About this article
Cite this article
Stuart, R.M., Romão, A.S., Pizzirani-Kleiner, A.A. et al. Culturable endophytic filamentous fungi from leaves of transgenic imidazolinone-tolerant sugarcane and its non-transgenic isolines. Arch Microbiol 192, 307–313 (2010). https://doi.org/10.1007/s00203-010-0557-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-010-0557-9