Abstract
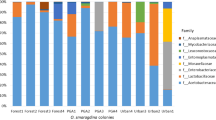
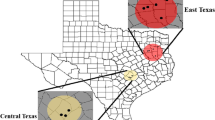
Social insects harbor diverse assemblages of bacterial microbes, which may play a crucial role in the success or failure of biological invasions. The invasive fire ant Solenopsis invicta (Formicidae, Hymenoptera) is a model system for understanding the dynamics of invasive social insects and their biological control. However, little is known about microbes as biotic factors influencing the success or failure of ant invasions. This pilot study is the first attempt to characterize and compare microbial communities associated with the introduced S. invicta and the native Solenopsis geminata in the USA. Using 16S amplicon 454 pyrosequencing, bacterial communities of workers, brood, and soil from nest walls were compared between neighboring S. invicta and S. geminata colonies at Brackenridge Field Laboratory, Austin, Texas, with the aim of identifying potential pathogenic, commensal, or mutualistic microbial associates. Two samples of S. geminata workers showed high counts of Spiroplasma bacteria, a known pathogen or mutualist of other insects. A subsequent analysis using PCR and sequencing confirmed the presence of Spiroplasma in additional colonies of both Solenopsis species. Wolbachia was found in one alate sample of S. geminata, while one brood sample of S. invicta had a high count of Lactococcus. As expected, ant samples from both species showed much lower microbial diversity than the surrounding soil. Both ant species had similar overall bacterial diversities, although little overlap in specific microbes. To properly characterize a single bacterial community associated with a Solenopsis ant sample, rarefaction analyses indicate that it is necessary to obtain 5,000–10,000 sequences. Overall, 16S amplicon 454 pyrosequencing appears to be a cost-effective approach to screen whole microbial diversity associated with invasive ant species.


Similar content being viewed by others
References
Allen GE, Buren WF (1974) Microsporidan and fungal diseases of Solenopsis invicta Buren in Brazil. J NY Entomol Soc 82:125–130
Bailey MT, Dowd SE, Parry NMA, Galley JD, Schauer DB, Lyte M (2010) Stressor exposure disrupts commensal microbial populations in the intestines and leads to increased colonization by Citrobacter rodentium. Infect Immun 78:1509–1519
Baird R, Woolfolk S, Watson CE (2007) Survey of bacterial and fungal associates of black/hybrid imported fire ants from mounds in Mississippi. Southeast Nat 6:615–632
Borm SV, Buschinger A, Boomsma JJ, Billen J (2002) Tetraponera ants have gut symbionts related to nitrogen-fixing root-nodule bacteria. Proc R Soc Lond B 269:2023–2027
Bouwma AM, Ahrens ME, DeHeer CJ, Shoemaker DD (2006) Distribution and prevalence of Wolbachia in introduced populations of the fire ant Solenopsis invicta. Insect Mol Biol 15:89–93
Briano J, Patterson RA (1994) The impact of the microsporidian disease, Thelohania solenopsae, on field populations of Solenopsis richteri in Argentina. In: Lenoir A et al (eds) Les insectes Sociaux 12th Congress of the International Union for the Study of Social Insects, Paris, Sorbonne, 21–27 August 1994. Université Paris Nord, Paris, p 116
Callaway TR, Dowd SE, Wolcott RD, Sun Y, Mcreynolds JL, Edrington TS, Byrd JA, Anderson RC, Krueger N, Nisbet DJ (2009) Evaluation of the bacterial diversity in cecal contents of laying hens fed various molting diets by using bacterial tag-encoded FLX amplicon pyrosequencing. Poult Sci 88:298–302
Clark TB (1984) Diversity of Spiroplasma host–parasite relationships. Isr J Med Sci 20:995–997
Cole RM, Tully JG, Popkin TJ, Bove JM (1973) Ultrastructure of the agent of citrus “stubborn” disease. Ann NY Acad Sci 222:471–493
de Souza DJ, Bezier A, Depoix D, Drezen JM, Lenoir A (2009) Blochmannia endosymbionts improve colony growth and immune defence in the ant Camponotus fellah. BMC Microbiol 9:29. doi:10.1186/1471-2180-9-29
Dowd SE, Sun Y, Secor PR, Rhoads DD, Wolcott BM, James GA, Wolcott RD (2008) Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol 8:43. doi:10.1186/1471-2180-8-43
Dowd SE, Zaragoza J, Rodriguez JR, Oliver MJ, Payton PR (2005) Windows.NET network distributed basic local alignment search toolkit (W.ND-BLAST). BMC Bioinform 6:93
Feener DH Jr, Brown BV (1992) Reduced foraging of Solenopsis geminata (Hymenoptera: Formicidae) in the presence of parasitic Pseudacteon spp. (Diptera: Phoridae). Ann Entomol Soc Am 85:80–84
Feener DH, Orr MR, Wackford K, Longo J, Benson WW, Gilbert LE (2008) Geographic variation in resource dominance—discovery in Brazilian ant communities. Ecology 89:1824–1836
Fierer N, Lauber CL, Zhou N, McDonald D, Costello EK, Knight R (2010) Forensic identification using skin bacterial communities. Proc Natl Acad Sci USA 107:6477–6481
Glancey BM, Vander Meer RK, Glover A, Lofgren CS, Vinson SB (1981) Filtration of microparticles from liquids ingested by the red imported fire ant Solenopsis invicta Buren. Insect Soc 28:395–401
Gontcharova V, Youn E, Wolcott RD, Hollister EB, Gentry TJ, Dowd SE (2010) Black box chimera check (B2C2): a Windows-based software for batch depletion of chimeras from bacterial 16S rRNA gene datasets. Open Microbiol J 4:47–52
Gunawan S, Tufts DM, Bextine BR (2008) Molecular identification of hemolymph-associated symbiotic bacteria in red imported fire ant larvae. Curr Microbiol 57:575–579
Hackett KJ, Clark TB (1989) The ecology of spiroplasmas. In: Whitcomb RF, Tully JG (eds) The mycoplasmas. Academic, New York, pp 113–200
Hamady M, Lozupone C, Knight R (2010) Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 4:17–27
Haselkorn TS, Markow TA, Moran NA (2009) Multiple introductions of the Spiroplasma bacterial endosymbiont into Drosophila. Mol Ecol 18:1294–1305
Jaenike J, Stahlhut J, Boelio L, Unckless R (2010) Association between Wolbachia and Spiroplasma within Drosophila neotestacea: an emerging symbiotic mutualism? Mol Ecol 19:414–425
Jaenike J, Unckless R, Cockburn S, Boelio L, Perlman S (2010) Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science 329:212–215
Jouvenaz DP, Allen GE, Banks WA, Wojcik DP (1977) A survey for pathogens of fire ants, Solenopsis spp., in the Southeastern United States. Fla Entomol 60:275–279
Jouvenaz DP, Ellis EA (1986) Vairimorpha invictae n. sp. (Microspora: Microsporida), a parasite of the red imported fire ant, Solenopsis invicta Buren (Hymenoptera: Formicidae). J Protozool 33:457–461
King JR, Tschinkel WR (2008) Experimental evidence that human impacts drive fire ant invasions and ecological change. Proc Natl Acad Sci USA 105:20339–20343
Knell JD, Allen GE, Hazard EI (1977) Light and electron microscope study of Thelohania solenopsae n. sp. (Microsporida: Protozoa) in the red imported fire ant, Solenopsis invicta. J Invertebr Pathol 29:192–200
Lee AH, Husseneder C, Hooper-Bui L (2008) Culture-independent identification of gut bacteria in fourth-instar red imported fire ant, Solenopsis invicta Buren, larvae. J Inverteb Pathol 98:20–33
Li HW, Medina F, Vinson SB, Coates CJ (2005) Isolation, characterization, and molecular identification of bacteria from the red imported fire ant (Solenopsis invicta) midgut. J Inverteb Pathol 89:203–209
Li W, Jaroszewski L, Godzik A (2001) Clustering of highly homologous sequences to reduce the size of large protein database. Bioinformatics 17:282–283
Liu Z, Lozupone C, Hamady M, Bushman FD, Knight R (2007) Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res 35:e120
Lozupone C, Hamady M, Kelley ST, Knight R (2007) Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73:1576–1585
Medina F, Li H, Vinson SB, Coates CJ (2009) Genetic transformation of midgut bacteria from the red imported fire ant (Solenopsis invicta). Curr Microbiol 58:478–82
Morrison LW (2000) Mechanisms of interspecific competition among an invasive and two native fire ants. Oikos 90:238–252
Mouches C, Bove JM, Albisetti J (1984) Pathogenicity of Spiroplasma apis and other spiroplasmas for honey-bees in southwestern France. Ann Microbiol Paris 135A:151–155
Oi DH, Pereira RM (1993) Ant behavior and microbial pathogens (Hymenoptera: Formicidae). Fla Entomol 76:63–73
Oi DH, Valles SM (2009) Fire ant control with entomopathogens in the USA. In: Hajek A et al (eds) Use of microbes for control and eradication of invasive arthropods. Springer Science+Business Media B.V, Dordercht, pp 237–257
Peloquin JJ, Greenberg L (2003) Identification of midgut bacteria from fourth instar red imported fire ant larvae, Solenopsis invicta Buren (Hymenoptera: Formicidae). J Agric Urban Entomol 20:157–164
Plowes RM, Dunn JG, Gilbert LE (2007) The urban fire ant paradox: native fire ants persist in an urban refuge while invasive fire ants dominate natural habitats. Biol Invasions 9:825–836
Porter SD, Williams DF, Patterson RS, Fowler HG (1997) Intercontinental differences in the abundance of Solenopsis fire ants (Hymenoptera: Formicidae): an escape from natural enemies? Environ Entomol 26:373–384
Price MN, Dehal PS, Arkin AP (2009) FastTree: computing large minimum-evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650
Regassa LB, Gasparich GE (2006) Spiroplasmas: evolutionary relationships and biodiversity. Front Biosci 11:2983–3002
Russell JA, Moreau CS, Goldman-Huertas B, Fujiwara M, Lohman DJ, Pierce NE (2009) Bacterial gut symbionts are tightly linked with the evolution of herbivory in ants. Proc Natl Acad Sci USA 106:21236–21241
Sánchez-Peña SR, Thorvilson HG (1992) Two fungi infecting red imported fire ant founding queens from Texas. Southwest Entomol 17:181–182
Sauer C, Stackebrandt E, Gadau J, Holldobler B, Gross R (2000) Systematic relationships and cospeciation of bacterial endosymbionts and their carpenter ant host species: proposal of the new taxon Candidatus Blochmannia gen. nov. Int J Syst Evol Microbiol 50:1877–1886
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Schluns H, Crozier RH (2009) Molecular and chemical immune defenses in ants (Hymenoptera: Formicidae. Myrmecological News 12:237–249
Sen R, Ishak HD, Estrada D, Dowd SE, Hong E, Mueller UG (2009) Generalized antifungal activity and 454-screening of Pseudonocardia and Amycolatopsis bacteria in nests of fungus-growing ants. Proc Natl Acad Sci USA 106:17805–17810
Shoemaker DD, Ahrens M, Sheill L, Mescher M, Keller L, Ross KG (2003) Distribution and prevalence of Wolbachia infections in native populations of the fire ant Solenopsis invicta (Hymenoptera: Formicidae). Environ Entomol 32:1329–1336
Smith DM, Snow DE, Rees E, Zischkau AM, Hanson JD, Wolcott RD, Sun Y, White J, Kumar S, Dowd SE (2010) Evaluation of the bacterial diversity of pressure ulcers using bTEFAP pyrosequencing. BMC Med Genomics 3:41
Stimac JL, Alves SB (1994) Ecology and biological control of fire ants. In: Rosen D et al (eds) Pest Management in the subtropics: biological control—a Florida perspective. Intercept Ltd., Andover, pp 353–380
Teixeira L, Ferreira A, Ashburner M (2008) The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol 6:e1000002
Tschinkel WR (1988) Distribution of the fire ants Solenopsis invicta and S. geminata (Hymenoptera: Formicidae) in northern Florida in relation to habitat and disturbance. Ann Entomol Soc Am 81:76–81
Tschinkel WR (2005) The fire ants. Harvard University Press, Cambridge, p 747
Tufts DM, Bextine B (2009) Identification of bacterial species in the hemolymph of queen Solenopsis invicta (Hymenoptera: Formicidae). Environ Entomol 38:1360–1364
Valles SM, Strong CA, Dang PM, Hunter WB, Pereira RM, Oi DH, Shapiro AM, Williams DF (2004) A picorna-like virus from the red imported fire ant, Solenopsis invicta: initial discovery, genome sequence, and characterization. Virology 328:151–157
Valles SM, Strong CA, Hashimoto Y (2007) A new positive-strand RNA virus with unique genome characteristics from the red imported fire ant, Solenopsis invicta. Virology 365:457–463
Vinson, SB, Sorensen, AA (1986) Imported fire ants: life history and impact. Texas Department of Agriculture, p 28
Williams DF, Knue GJ, Becnel JJ (1998) Discovery of Thelohania solenopsae from the red imported fire ant, Solenopsis invicta, in the United States. J Inverteb Pathol 71:175–176
Wojcik DP, Allen CR, Brenner RJ, Forys EA, Jouvenaz DP, Lutz RS (2001) Red imported fire ants: impact on biodiversity. Am Entomol 47:16–23
Xie J, Vilchez I, Mateos M (2010) Spiroplasma bacteria enhance survival of Drosophila hydei attacked by the parasitic wasp Leptopilina heterotoma. PLoS ONE 5:e12149. doi:10.1371/journal.pone.0012149
Acknowledgments
We thank Edward LeBrun for assisting in the collection of samples. Larry Gilbert, Edward LeBrun, and Jon Seal provided constructive comments on the manuscript. The work was funded by the Lee and Ramona Bass Foundation, the Helen C. Kleberg & Robert J. Kleberg Foundation, NSF Grant DEB-0639879, and by the W.M. Wheeler Lost-Pines endowment from the University of Texas at Austin.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary Figure 1
Rarefaction curves for each nest for using 1% (species level) and 3% (generic level) OTU designations (DOC 1312 kb)
Supplementary Figure 2
UniFrac weighted principal coordinate analyses (PCoA) for all S. geminata and S. invicta ant samples. The arrow indicates two S. geminata workers (A4 and A9) which have overlapping symbols due to their high numbers of Spiroplasma sequences. The bacterial communities between S. geminata and S. invicta have some overlap and are not distinctly separate (DOC 308 kb)
Supplementary Table 1
BLAST results according to its nearest genus (matched at 100–75% identity) for all 454-generated sequences and analyzed separately for each sample providing relative abundance percentages (XLS 222 kb)
Supplementary Table 2
Conservative BLAST results for all 454 generated sequence data (XLS 416 kb)
Supplementary Table 3
Unique sequence ID’s, BLAST hit, and identity scores for all 454-generated sequences (XLS 9146 kb)
Rights and permissions
About this article
Cite this article
Ishak, H.D., Plowes, R., Sen, R. et al. Bacterial Diversity in Solenopsis invicta and Solenopsis geminata Ant Colonies Characterized by 16S amplicon 454 Pyrosequencing. Microb Ecol 61, 821–831 (2011). https://doi.org/10.1007/s00248-010-9793-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-010-9793-4