Abstract
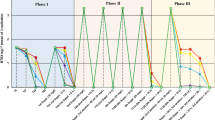
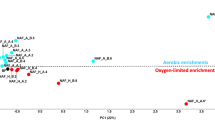
A set of microcosm experiments was performed to assess different bioremediation strategies, i.e., biostimulation and bioaugmentation, for groundwater contaminated with chlorobenzenes. The biodegradative potential was stimulated either by the supply of electron acceptors (air, (NO −3 ), to increase the activity of the indigenous bacterial community, or by the addition of aerobic chlorobenzene-degrading bacteria (Pseudomonas putida GJ31, Pseudomonas aeruginosa RHOl, Pseudomonas putida F1ΔCC). Experiments were performed with natural groundwater of the aquifer of Bitterfeld, which had been contaminated with 1,2-dichlorobenzene (1,2-DCB), 1,4-dichlorobenzene (1,4-DCB), and chlorobenzene (CB). The microcosms consisted of airtight glass bottles with 800 mL of natural groundwater and were incubated under in situ temperature (13°C). Behavior of the introduced strains within the indigenous bacterial community was monitored by fluorescent in situ hybridization (FISH) with species-specific oligonucleotides. Dynamics of the indigenous community and the introduced strains within the microcosms were followed by single-strand conformation polymorphism (SSCP) analysis of 16S rDNA amplicons obtained from total DNA of the microbial community. An indigenous biodegradation potential under aerobic as well as anaerobic denitrifying conditions was observed accompanied by fast and specific changes in the natural bacterial community composition. Augmentation with P. aeruginosa RHO1 did not enhance bio-degradation. In contrast, both P. putida GJ31 as well as P. putida F1ΔCC were capable of growing in groundwater, even in the presence of the natural microbial community, and thereby stimulating chlorobenzene depletion. P. putida GJ31 disappeared when the xenobiotics were depleted and P. putida F1ΔCC persisted even in the absence of CB. Detailed statistical analyses revealed that community dynamics of the groundwater microbiota were highly reproducible but specific to the introduced strain, its inoculum size, and the imposed physicochemical conditions. These findings could contribute to the design of better in situ bioremediation strategies for contaminated groundwater.
Similar content being viewed by others
References
Adrian L, Szewzyk U, Wecke J, Gorisch H (2000) Bacterial dehalorespiration with chlorinated benzenes. Nature 408: 580–583
Amann R, Ludwig W, Schleifer K-H (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microb Rev 59:143–169
Bassam BJ, Caetano AG, Gresshoff PM (1991) Fast and sensitive silver staining of DNA in Polyacrylamid gels. Anal Biochem 80:81–84
Boon N, Goris J, De Vos P, Verstraete W, Top EM (2000) Bioaugmentation of activated sludge by an indigenous 3-chloroaniline-degrading Comamonas testosteroni strain, I2gfp. Appl Environ Microbiol 66:2906–2913
Bradford MM (1976) A rapid and sensitive method for the quantitation of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Bragg JR, Prince RC, Harner EJ, Atlas RM (1994) Effectiveness of bioremediation for the Exxon Valdez oil spill. Nature 368:413–418
Brunsbach TR, Reineke W (1994) Degradation of chlorobenzenes in soil slurry by a specialized organism. Appl Microbiol Biotechnol 42:415–420
Bulman TL, Newland M (1993) In situ bioventing of a diesel fuel spill. Hydrol Sei 8:297–308
Cho JC, Kim SJ (2000) Computer-assisted PCR-single-strand conformation polymorphism analysis for assessing shift in soil bacterial community structure during bioremidiational treatments. World J Microbiol Biotechnol 16:231–235
De Leij F, Sutton E, Whipps J, Fenlon J, Lynch J (1995) Impact of field release of genetically modified Pseudomonas fluoresceins on indigenous microbial populations of wheat. App Environ Microbiol 61:3443–3453
Delbès C, Moletta R, Godon J (2000) Monitoring of activity dynamics of an anaerobic digester bacterial community using 16S rRNA polymerase chain reaction-single-strand conformation polymorphism analysis. Environ Microbiol 2:506–515
Dorn E, Hellwig M, Reineke W, Knackmuss H-J (1974) Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch Microbiol 99:61–70
Duba AG, Jackson KJ, Jovanovich MC, Knapp RB, Taylor RT (1996) TCE remediation using in situ, resting-state bioaugmentation. Environ Sei Technol 30:1982–1989
DuTeau NM, Rogers JD, Bartholomay CT, Reardon KF (1998) Species-specific oligonucleotides for enumeration of Pseudomonas putida Fl, Burkholderia sp. strain JS150, and Bacillus subtilis ATCC 7003 in biodegradation experiments. Appl Environ Microbiol 64:4994–4999
Dybas MJ, Barcelona M, Bezborodnikov S, Davies S, Forney L, Heuer H, Kawka O, Mayotte T, Sepülveda-Torres L, Smalla K, Sneathen M, Tiedje J, Voice T, Wiggert DC, Witt DC, Criddle CS (1998) Pilot-scale evaluation of bioaugmentation for in-situ remediation of carbon tetrachloride-contaminated aquifer. Environ Sei Technol 32:3598–3611
Fuhrman JA, Comeau DE, Hagström Å, Chan AM (1988) Extraction from natural planktonic microorganisms of DNA suitable for molecular biological studies. Appl Environ Microbiol 54:1426–1429
Gerritse J, Drzyzga O, Kloetstra G, Keijmel M, Wiersum LP, Hutson R, Collins MD, Gottschal JC (1999) Influence of different electron donors and accepters on dehalorespiration of tetrachloroethene by Desulfitobacterium frappieri TCE1. Appl Environ Microbiol 65:5212–5221
Haigler B, Nishino S, Spain J (1988) Degradation of 1,2-dichlorobenzene by a Pseudomonas sp. Appl Environ Microbiol 54:294–301
Holliger C, Schumacher W (1994) Reductive dehalogenation as a respiratory process. Antonie van Leeuwenhoek 66:239–246
Lehning A (1998) PhD thesis. Technical University, Braunschweig, Germany
Mars AE, Kasberg T, Kaschabek SR, van Agteren MH, Janssen DB, Reineke W (1997) Microbial degradation of chloroaromatics: use of the mete-cleavage pathway for mineralization of chlorobenzene. J Bacteriol 179:4530–4537
Miethling R, Karlson U (1996) Accelerated mineralization of pentachlorophenol in soil upon inoculation with Mycobacterium chlorophenolicum PCP1 and Sphingomonas chloro-phenolica RA2. Appl Environ Microbiol 62:4361–4366
Neumann A, Wohlfarth G, Diekert G (1995) Properties of tetrachloroethene and trichloroethene dehalogenase of Dehalospirillum multivorans. Arch Microbiol 163:276–281
Nishino SF, Spain JC, Belcher LA, Litchfield CD (1992) Chlorobenzene degradation by bacteria isolated from contaminated groundwater. Appl Environ Microbiol 58:1719–1726
Oltmanns R, Rast H, Reineke W (1988) Degradation of 1,4-dichlorobenzene by enriched and constructed bacteria. Appl Microbiol Biotechnol 28:609–616
Pieper DH, Stadler-Fritzsche K, Engesser K-H, Knackmuss H-J (1993) Metabolism of 2-chloro-4-methylphenoxyacetate by Alcaligenes eutrophus JMP 134. Arch Microbiol 160:169–178
Popp P, Moder M (1997) Organische Analytik. UFZ-Bericht, Leipzig
Ramanand K, Balba MT, Duffy J (1993) Reductive dehalo-genation of chlorinated benzenes and toluenes under methanogenic conditions. Appl Environ Microbiol 59:3266–3272
Ravatn R, Zehnder A, van der Meer J (1998) Low-frequency horizontal transfer of an element containing the chlorocatechol degradation genes from Pseudomonas sp. strain B13 to Pseudomonas putida Fl and to indigenous bacteria in laboratory-scale activated-sludge mircocosms. Appl Environ Microbiol 64:2126–2132
Reineke W, Knackmuss H-J (1984) Microbial metabolism of haloaromatics: Isolation and properties of a chlorobenzene-degrading bacterium. Appl Environ Microbiol 47:395–402
Relman DA (1993) Universal Bacterial 16S rDNA Amplification and Sequencing. American Society for Microbiology, Washington, DC
Ruske R, Hübner J, Böhme 0, Falke P (1997) Ergebnisse der geologisch-hydrogeologischen-geotechnischen Standorter kundung. UFZ-Bericht, Leipzig
Salanitro JP, Johnson PC, Spinnler GE, Maner PM, Wisniewski HL, Bruce C (2000) Field scale demonstration of enhanced MTBE bioremediation through aquifer bioaugmentation and oxygenation. Environ Sei Technol 34:4152–4162
Schwieger F, Tebbe CC (1998) A new approach to utilize PCR-single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl Environ Microbiol 64:4870–4876
Sneath PHA, Sokal RR (1973) Numerical Taxonomy. Freeman, San Francisco
Stoffels M, Amann R, Ludwig W, Hekmat D, Schleifer K (1998) Bacterial community dynamics during start-up of a trickle-bed bioreactor degrading aromatic compounds. Appl Environ Microbiol 64:930–939
Tchelet R, Meckenstock R, Steinle P, van der Meer JR (1999) Population dynamics of an introduced bacterium degrading chlorinated benzenes in a soil column and in sewage sludge. Biodegradation 10:113–125
Tranvik L, Höfle M (1987) Bacterial growth in mixed cultures on dissolved organic carbon from humic and clear waters. Appl Environ Microbiol 53:482–488
Utkin I, Dalton DD, Wiegel J (1995) Specificity of reductive dehalogenation of substituted ortho-chlorophenols by Desulfitobacterium dehalogenans JW/IU-DC1. Appl Environ Microbiol 61:346–351
van de Pas BA, Smidt H, Hagen WR, van der Oost J, Schraa G, Stams AJM, de Vos WM (1999) Purification and molecular characterization of orho-chlorophenol reductive dehalogenase, a key enzyme of halorespiration in Desulfitobacterium dehalogenans. J Biol Chem 274:20287–20292
van der Meer J, Eggen R, Zehnder A, de Vos W (1991) Sequence analysis of the Pseudomonas sp. strain P51 tcb gene cluster, which encodes metabolism of chlorinated catechols: evidence for specialization of catechol 1,2-dioxygenases for chlorinated substrates. J Bacteriol 173:2425–2434
van der Meer J, Roelofson W, Schraa G, Zehnder A (1987) Degradation of low concentrations of dichlorobenzenes and 1,2,4-trichlorobenzene by Pseudomonas sp. strain P51 in nonsterile soil columns. FEMS Microbiol Ecol 45:333–341
van der Meer J, Werklen C, Nishino S, Spain J (1998) Evolution of a pathway for chlorobenzene metabolism leads to natural attenuation in contaminated groundwater. Appl Environ Microbiol 64:4185–4193
Van Limbergen H, Topp EM, Verstraete W (1998) Bioaugmentation in activated sludge: current features and future perspective. Appl Microbiol Biotechnol 50:16–23
Watanabe K, Yamamoto S, Hino S, Harayama S (1998) Population dynamics of phenol-degrading bacteria in activated sludge determined by gyrB-targeted quantitative PCR. Appl Environ Microbiol 64:1203–1209
Weinbauer MG, Beckmann C, Höfle MG (1998) Utility of green fluorescent nucleic acid dyes and aluminum oxide membrane filters for rapid epifluorescence enumeration of soil and sediment bacteria. Appl Environ Microbiol 64:5000–5003
Weiß H, Kopinke F, Popp P, Wünsche L (1998) In situ remediation research in a complexly contaminated aquifer: the SAFIRA test site at Bitterfeld. Germany-NATO/CCMS Pilot Study Special Session 229:84–91
Wikstrom P, Hagglund L, Forsman M (2000) Structure of a natural microbial community in a nitroaromatic contaminated groundwater is altered during biodégradation of extrinsic, but not intrinsic substrates. Microb Ecol 39:203–210
Zylstra G, McCombie W, Gibson D, Finette B (1988) Toluene degradation by Pseudomonas putida F1: genetic organization of the tod Operon. Appl Environ Microbiol 54:1498–1503
Author information
Authors and Affiliations
Corresponding author
Additional information
Online publication: 6 June 2003
Rights and permissions
About this article
Cite this article
Wenderoth, D.F., Rosenbrock, P., Abraham, W.R. et al. Bacterial community dynamics during biostimulation and bioaugmentation experiments aiming at chlorobenzene degradation in groundwater. Microb Ecol 46, 161–176 (2003). https://doi.org/10.1007/s00248-003-2005-8
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s00248-003-2005-8