Abstract.
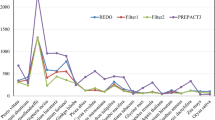
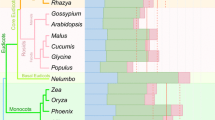
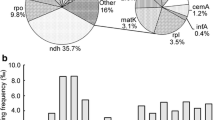
Transcripts of land plant chloroplast genomes undergo C-to-U RNA editing. Systematic search disclosed 31 editing sites in tobacco, 27 in maize, and 21 in rice. Based on these identified sites, potential editing sites have been predicted in the transcripts from four angiosperm chloroplast genomes which have been completely sequenced. Most RNA editing events occur in internal codons, which result in amino-acid substitutions. The initiation codon AUG was found to be created from ACG by RNA editing in the transcripts from rpl2, psbL, and ndhD genes. Comparison of editing patterns raises a possibility that many editing sites were acquired in the evolution of angiosperms.
Similar content being viewed by others
Author information
Authors and Affiliations
Additional information
Received: 18 January 2001 / Accepted: 28 February 2001
Rights and permissions
About this article
Cite this article
Tsudzuki, T., Wakasugi, T. & Sugiura, M. Comparative Analysis of RNA Editing Sites in Higher Plant Chloroplasts. J Mol Evol 53, 327–332 (2001). https://doi.org/10.1007/s002390010222
Issue Date:
DOI: https://doi.org/10.1007/s002390010222