Abstract
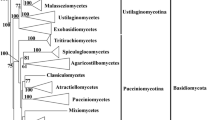
We have used three independent phylogenomic approaches (concatenated alignments, single-, and multi-gene supertrees) to reconstruct the fungal tree of life (FTOL) using publicly available fungal genomes. This is the first time multi-gene families have been used in fungal supertree reconstruction and permits us to use up to 66% of the 1,001,217 genes in our fungal database. Our analyses show that different phylogenomic datasets derived from varying clustering criteria and alignment orientation do not have a major effect on phylogenomic supertree reconstruction. Overall the resultant phylogenomic trees are relatively congruent with one another and successfully recover the major fungal phyla, subphyla and classes. We find that where incongruences do occur, the inferences are usually poorly supported. Within the Ascomycota phylum, our phylogenies reconstruct monophyletic Saccharomycotina and Pezizomycotina subphyla clades and infer a sister group relationship between these to the exclusion of the Taphrinomycotina. Within the Pezizomycotina subphylum, all three phylogenies infer a sister group relationship between the Leotiomycetes and Sordariomycetes classes. However, there is conflict regarding the relationships with the Dothideomycetes and Eurotiomycetes classes. Within the Basidiomycota phylum, supertrees derived from single- and multi-gene families infer a sister group relationship between the Pucciniomycotina and Agaricomycotina subphyla while the concatenated phylogeny infers a poorly supported relationship between the Agaricomycotina and Ustilagomycotina. The reconstruction of a robust FTOL is important for future fungal comparative analyses. We illustrate this point by performing a preliminary investigation into the phyletic distribution of yeast prion-like proteins in the fungal kingdom.





Similar content being viewed by others
References
Alberti S, Halfmann R, King O, Kapila A, Lindquist S (2009) A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137:146–158
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res (Online) 25:3389–3402
Archie JW (1989) A randomization test for phylogenetic information in systematic data. Syst Zool 38:251–278
Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF (2000) A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290:972–977
Barrett M, Donoghue MJ, Sober E (1991) Against consensus. Syst Zool 40:486–493
Baum BR (1992) Combining trees as a way of combining data sets for phylogenetic inference, and the desirability of combining gene trees. Taxon 41:3–10
Begerow D, John B, Oberwinkler F (2004) Evolutionary relationships among beta-tubulin gene sequences of basidiomycetous fungi. Mycol Res 108:1257–1263
Blackwell M, Hibbett DS, Taylor JW, Spatafora JW (2006) Research Coordination Networks: a phylogeny for kingdom Fungi (Deep Hypha). Mycologia 98:829–837
Brachmann A, Baxa U, Wickner RB (2005) Prion generation in vitro: amyloid of Ure2p is infectious. EMBO J 24:3082–3092
Bullerwell CE, Forget L, Lang BF (2003) Evolution of monoblepharidalean fungi based on complete mitochondrial genome sequences. Nucleic Acids Res 31:1614–1623
Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, Agrafioti I, Arnaud MB, Bates S, Brown AJ, Brunke S, Costanzo MC, Fitzpatrick DA, de Groot PW, Harris D, Hoyer LL, Hube B, Klis FM, Kodira C, Lennard N, Logue ME, Martin R, Neiman AM, Nikolaou E, Quail MA, Quinn J, Santos MC, Schmitzberger FF, Sherlock G, Shah P, Silverstein KA, Skrzypek MS, Soll D, Staggs R, Stansfield I, Stumpf MP, Sudbery PE, Srikantha T, Zeng Q, Berman J, Berriman M, Heitman J, Gow NA, Lorenz MC, Birren BW, Kellis M, Cuomo CA (2009) Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459(7247):657–662
Byrne KP, Wolfe KH (2005) The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res 15:1456–1461
Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552
Creevey CJ, McInerney JO (2005) Clann: investigating phylogenetic information through supertree analyses. Bioinformatics 21:390
Creevey CJ, Fitzpatrick DA, Philip GK, Kinsella RJ, O’Connell MJ, Pentony MM, Travers SA, Wilkinson M, McInerney JO (2004) Does a tree-like phylogeny only exist at the tips in the prokaryotes? Proc Biol Sci 271:2551–2558
Dagan T, Martin W (2006) The tree of one percent. Genome Biol 7:118
De Schutter K, Lin YC, Tiels P, Van Hecke A, Glinka S, Weber-Lehmann J, RouzÈ P, Van de Peer Y, Callewaert N (2009) Genome sequence of the recombinant protein production host Pichia pastoris. Nat Biotechnol 27:561–566
Delsuc F, Brinkmann H, Philippe H (2005) Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet 6:361–375
Diezmann S, Cox CJ, Schonian G, Vilgalys RJ, Mitchell TG (2004) Phylogeny and evolution of medical species of Candida and related taxa: a multigenic analysis. J Clin Microbiol 42:5624–5635
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792
Eernisse D, Kluge A (1993) Taxonomic congruence versus total evidence, and amniote phylogeny inferred from fossils, molecules, and morphology. Mol Biol Evol 10:1170–1195
Enright AJ, Van Dongen S, Ouzounis CA (2002) An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30:1575
Faith DP, Cranston PS (1991) Could a cladogram this short have arisen by chance alone? On permutation tests for cladistic structure. Cladistics 7:1–28
Faris JD, Zhang Z, Lu H, Lu S, Reddy L, Cloutier S, Fellers JP, Meinhardt SW, Rasmussen JB, Xu SS, Oliver RP, Simons KJ, Friesen TL (2010) A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proc Natl Acad Sci USA 107:13544–13549
Fitzpatrick DA, Logue ME, Stajich JE, Butler G (2006) A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol 6:99
Fitzpatrick DA, Logue ME, Butler G (2008) Evidence of recent interkingdom horizontal gene transfer between bacteria and Candida parapsilosis. BMC Evol Biol 8:181
Fitzpatrick DA, O’Gaora P, Byrne KP, Butler G (2010) Analysis of gene evolution and metabolic pathways using the Candida Gene Order Browser. BMC Genomics 11:290
Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Basturkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, D’Enfert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Penalva MA, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren BW (2005a) Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105–1115
Galagan JE, Henn MR, Ma LJ, Cuomo CA, Birren B (2005b) Genomics of the fungal kingdom: insights into eukaryotic biology. Genome Res 15:1620–1631
Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG (1996) Life with 6000 genes. Science 274(546):563–567
Guarro J, Gene J, Stchigel AM (1999) Developments in fungal taxonomy. Clin Microbiol Rev 12:454–500
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Harrison LB, Yu Z, Stajich JE, Dietrich FS, Harrison PM (2007) Evolution of budding yeast prion-determinant sequences across diverse fungi. J Mol Biol 368:273–282
Hawksworth DL (1991) The fungal dimension of biodiversity: magnitude, significance, and conservation. Mycol Res 95:641–655
Hawksworth DL (2001) The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycol Res 109:1422–1432
Heckman DS, Geiser DM, Eidell BR, Stauffer RL, Kardos NL, Hedges SB (2001) Molecular evidence for the early colonization of land by fungi and plants. Science 293:1129–1133
Hibbett DS (2006) A phylogenetic overview of the Agaricomycotina. Mycologia 98:917–925
Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, Eriksson OE, Huhndorf S, James T, Kirk PM, Lücking R (2007) A higher-level phylogenetic classification of the Fungi. Mycol Res 111:509–547
Holton TA, Pisani D (2010) Deep genomic-scale analyses of the metazoa reject Coelomata: evidence from single- and multigene families analyzed under a supertree and supermatrix paradigm. Genome Biol Evol 2:310–324
James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, Lumbsch HT, Rauhut A, Reeb V, Arnold AE, Amtoft A, Stajich JE, Hosaka K, Sung GH, Johnson D, O’Rourke B, Crockett M, Binder M, Curtis JM, Slot JC, Wang Z, Wilson AW, Schussler A, Longcore JE, O’Donnell K, Mozley-Standridge S, Porter D, Letcher PM, Powell MJ, Taylor JW, White MM, Griffith GW, Davies DR, Humber RA, Morton JB, Sugiyama J, Rossman AY, Rogers JD, Pfister DH, Hewitt D, Hansen K, Hambleton S, Shoemaker RA, Kohlmeyer J, Volkmann-Kohlmeyer B, Spotts RA, Serdani M, Crous PW, Hughes KW, Matsuura K, Langer E, Langer G, Untereiner WA, Lucking R, Budel B, Geiser DM, Aptroot A, Diederich P, Schmitt I, Schultz M, Yahr R, Hibbett DS, Lutzoni F, McLaughlin DJ, Spatafora JW, Vilgalys R (2006a) Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443:818–822
James TY, Letcher PM, Longcore JE, Mozley-Standridge SE, Porter D, Powell MJ, Griffith GW, Vilgalys R (2006b) A molecular phylogeny of the flagellated fungi (Chytridiomycota) and description of a new phylum (Blastocladiomycota). Mycologia 98:860–871
Jeffries TW, Grigoriev IV, Grimwood J, Laplaza JM, Aerts A, Salamov A, Schmutz J, Lindquist E, Dehal P, Shapiro H (2007) Genome sequence of the lignocellulose-bioconverting and xylose-fermenting yeast Pichia stipitis. Nat Biotechnol 25:319–326
Jung PP, Friedrich A, Souciet JL, Louis V, Potier S, de Montigny J, Schacherer J (2010) Complete mitochondrial genome sequence of the yeast Pichia farinosa and comparative analysis of closely related species. Curr Genet 56:507–515
Keane TM, Naughton TJ, McInerney JO (2004) ModelGenerator: amino acid and nucleotide substitution model selection. National University of Ireland. http://bioinf.nuim.i.e/software/modelgenerator
King CY, Diaz-Avalos R (2004) Protein-only transmission of three yeast prion strains. Nature 428:319–323
Kluge AG (1989) A concern for evidence and a phylogenetic hypothesis of relationships among Epicrates (Boidae, Serpentes). Syst Biol 38:7–25
Kuramae EE, Robert V, Snel B, Weifl M, Boekhout T (2006) Phylogenomics reveal a robust fungal tree of life. FEMS Yeast Res 6:1213–1220
Landan G, Graur D (2007) Heads or tails: a simple reliability check for multiple sequence alignments. Mol Biol Evol 24:1380–1383
Lartillot N, Philippe H (2004) A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol 21:1095–1109
Lartillot N, Philippe H (2008) Improvement of molecular phylogenetic inference and the phylogeny of Bilateria. Philos Trans R Soc Lond B Biol Sci 363:1463–1472
Lartillot N, Brinkmann H, Philippe H (2007) Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol Biol 7(Suppl 1):S4
Lindquist S (2009) Protein folding sculpting evolutionary change. Cold Spring Harb Symp Quant Biol 74:103–108
Liu YJ, Hodson MC, Hall BD (2006) Loss of the flagellum happened only once in the fungal lineage: phylogenetic structure of kingdom Fungi inferred from RNA polymerase II subunit genes. BMC Evol Biol 6:74
Liu Y, Leigh JW, Brinkmann H, Cushion MT, Rodriguez-Ezpeleta N, Philippe H, Lang BF (2009) Phylogenomic analyses support the monophyly of Taphrinomycotina, including Schizosaccharomyces fission yeasts. Mol Biol Evol 26:27–34
Lumbsch HT, Schmitt I, Lindemuth R, Miller A, Mangold A, Fernandez F, Huhndorf S (2005) Performance of four ribosomal DNA regions to infer higher-level phylogenetic relationships of inoperculate euascomycetes (Leotiomyceta). Mol Phylogenet Evol 34:512–524
Lutzoni F, Kauff F, Cox CJ, McLaughlin D, Celio G, Dentinger B, Padamsee M, Hibbett D, James TY, Baloch E, Grube M, Reeb V, Hofstetter V, Schoch C, Arnold AE, Miadlikowska J, Spatafora J, Johnson D, Hambleton S, Crockett M, Shoemaker R, Sung G, Lucking R, Lumbsch T, O’Donnell K, Binder M, Diederich P, Ertz D, Gueidan C, Hansen K, Harris R, Hosaka K, Lim Y, Matheny B, Nishida H, Pfister D, Rogers J, Rossman A, Schmitt I, Sipman H, Stone J, Sugiyama J, Yahr R, Vilgalys R (2004) Assembling the fungal tree of life: progress, classification, and evolution of subcellular traits. Am J Bot 91:1446–1480
Marcet-Houben M, Gabaldon T (2009) The tree versus the forest: the fungal tree of life and the topological diversity within the yeast phylome. PLoS One 4:e4357
McKintosh E, Tabrizi SJ, Collinge J (2003) Prion diseases. J Neurovirol 9:183–193
Mullan LJ (2002) Multiple sequence alignment—the gateway to further analysis. Brief Bioinform 3:303–305
Page RD (1998) GeneTree: comparing gene and species phylogenies using reconciled trees. Bioinformatics 14:819–820
Patel BK, Liebman SW (2007) “Prion-proof” for [PIN+]: infection with in vitro-made amyloid aggregates of Rnq1p-(132–405) induces [PIN+]. J Mol Biol 365:773–782
Peterson SW (2008) Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia 100:205–226
Philippe H, Snell EA, Bapteste E, Lopez P, Holland PW, Casane D (2004) Phylogenomics of eukaryotes: impact of missing data on large alignments. Mol Biol Evol 21:1740–1752
Pisani D, Cotton JA, McInerney JO (2007) Supertrees disentangle the chimerical origin of eukaryotic genomes. Mol Biol Evol 24:1752–1760
Prusiner SB (1982) Novel proteinaceous infectious particles cause scrapie. Science 216:136–144
Ragan MA (1992) Matrix representation in reconstructing phylogenetic relationships among the eukaryotes. Biosystems 28:47–55
Robbertse B, Reeves JB, Schoch CL, Spatafora JW (2006) A phylogenomic analysis of the Ascomycota. Fungal Genet Biol 43(10):715–725
Saupe SJ (2007) A short history of small s: a prion of the fungus Podospora anserina. Prion 1:110–115
Scannell DR, Byrne KP, Gordon JL, Wong S, Wolfe KH (2006) Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature 440:341–345
Scannell DR, Frank AC, Conant GC, Byrne KP, Woolfit M, Wolfe KH (2007) Independent sorting-out of thousands of duplicated gene pairs in two yeast species descended from a whole-genome duplication. Proc Natl Acad Sci USA 104:8397–8402
Schoch CL, Sung GH, Lopez-Giraldez F, Townsend JP, Miadlikowska J, Hofstetter V, Robbertse B, Matheny PB, Kauff F, Wang Z (2009) The Ascomycota tree of life: a phylum-wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Syst Biol 58:224
Si K, Choi YB, White-Grindley E, Majumdar A, Kandel ER (2010) Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell 140:421–435
Slowinski JB, Page RD (1999) How should species phylogenies be inferred from sequence data? Syst Biol 48:814–825
Steenkamp ET, Wright J, Baldauf SL (2006) The protistan origins of animals and fungi. Mol Biol Evol 23:93–106
Suh SO, Blackwell M, Kurtzman CP, Lachance MA (2006) Phylogenetics of Saccharomycetales, the ascomycete yeasts. Mycologia 98:1006–1017
Tanaka M, Chien P, Naber N, Cooke R, Weissman JS (2004) Conformational variations in an infectious protein determine prion strain differences. Nature 428:323–328
Wehe A, Bansal MS, Burleigh JG, Eulenstein O (2008) DupTree: a program for large-scale phylogenetic analyses using gene tree parsimony. Bioinformatics 24:1540–1541
Wickner RB (1994) [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 264:566–569
Wickner RB, Shewmaker F, Edskes H, Kryndushkin D, Nemecek J, McGlinchey R, Bateman D, Winchester CL (2010) Prion amyloid structure explains templating: how proteins can be genes. FEMS Yeast Res 10:980–991
Wong KM, Suchard MA, Huelsenbeck JP (2008) Alignment uncertainty and genomic analysis. Science 319:473–476
Yuan X, Xiao S, Taylor TN (2005) Lichen-like symbiosis 600 million years ago. Science 308:1017–1020
Acknowledgments
We wish to acknowledge the financial support of the Irish Health Research Board (HRB). We also wish to acknowledge the SFI/HEA Irish Centre for High-End Computing (ICHEC) for the provision of computational facilities and support. EMMT was funded through a SFI-UREKA summer school and would like to thank Dr James McInerney and Dr Davide Pisani. He also wishes to thank his academic advisor Prof. Silvia Restrepo.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic Supplementary material.
Rights and permissions
About this article
Cite this article
Medina, E.M., Jones, G.W. & Fitzpatrick, D.A. Reconstructing the Fungal Tree of Life Using Phylogenomics and a Preliminary Investigation of the Distribution of Yeast Prion-Like Proteins in the Fungal Kingdom. J Mol Evol 73, 116–133 (2011). https://doi.org/10.1007/s00239-011-9461-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-011-9461-4