Abstract
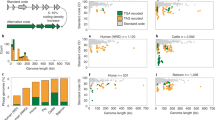
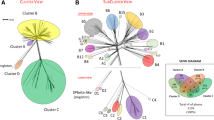
The size and diversity of bacteriophage populations require methodologies to quantitatively study the landscape of phage differences. Statistical approaches are confronted with small genome sizes forbidding significant single-phage analysis, and comparative methods analyzing full phage genomes represent an alternative but they are of difficult interpretation due to lateral gene transfer, which creates a mosaic spectrum of related phage species. Based on a large-scale codon bias analysis of 116 DNA phages hosted by 11 translationally biased bacteria belonging to different phylogenetic families, we observe that phage genomes are almost always under codon selective pressure imposed by translationally biased hosts, and we propose a classification of phages with translationally biased hosts which is based on adaptation patterns. We introduce a computational method for comparing phages sharing homologous proteins, possibly accepted by different hosts. We observe that throughout phages, independently from the host, capsid genes appear to be the most affected by host translational bias. For coliphages, genes involved in virion morphogenesis, host interaction and ssDNA binding are also affected by adaptive pressure. Adaptation affects long and small phages in a significant way. We analyze in more detail the Microviridae phage space to illustrate the potentiality of the approach. The small number of directions in adaptation observed in phages grouped around ϕX174 is discussed in the light of functional bias. The adaptation analysis of the set of Microviridae phages defined around ϕMH2K shows that phage classification based on adaptation does not reflect bacterial phylogeny.


Similar content being viewed by others
References
Akashi H (2001) Gene expression and molecular evolution. Curr Opin Genet Dev 11:660–666
Bailly-Bechet M, Vergassola M, Rocha E (2007) Causes for the intriguing presence of tRNAs in phages. Genome Res 17(10):1486–1495
Bennetzen JL, Hall BD (1982) Codon selection in yeast. J Biol Chem 257:3026–3031
Beres SB, Sylva GL, Barbian KD et al (2002) Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc Natl Acad Sci USA 99(15):10078–10083
Brentlinger KL, Hafenstein S, Novak CR, Fane BA, Borgon R, McKenna R, Agbandje-McKenna M (2002) Microviridae, a family divided: isolation, characterization, and genome sequence of ϕMH2K, a bacteriophage of the obligate intracellular parasitic bacterium Bdellovibrio bacteriovorus. J Bacteriol 184(4):1089–1094
Carbone A (2006) Computational prediction of genomic functional cores specific to different microbes. J Mol Evol 63(6):733–746
Carbone A, Madden R (2005) Insights on the evolution of metabolic networks of unicellular translationally biased organisms from transcriptomic data and sequence analysis. J Mol Evol 61:456–469
Carbone A, Zinovyev A, Képès F (2003) Codon Adaptation Index as a measure of dominating codon bias. Bioinformatics 19:2005–2015
Carbone A, Képès F, Zinovyev A (2005) Codon bias signatures, organization of microorganisms in codon space and lifestyle. Mol Biol Evol 22:547–561
Coetzee JN (1987) Bacteriophage taxonomie. In: Goyal SM, Gerba CP, Bitton G (eds) Phage ecology. Wiley and Sons Interscience, New York, pp 45–86
Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16(22):10881–10890
Cowe E, Sharp PM (1991) Molecular evolution of bacteriophages: discrete patterns of codon usage in T4 genes are related to the time of gene expression. J Mol Evol 33(1):13–22
Fiers W, Sinsheimer RL (1962) The structure of the DNA of bacteriophage ϕX174. III. Ultracentrifuge evidence for a ring structure. J Mol Biol, 5:424–434
Filée J, Forterre P, Laurent J (2003) The role played by viruses in the evolution of their hosts: a view based on informational protein phylogenies. Res Microbiol 154:237–243
Gomathi NS, Sameer H, Kumar V et al (2007) In silico analysis of mycobacteriophage Che 12 genome: characterisation of genes required to lysogenise Mycobacterium tuberculosis. Comput Biol Chem 31(2):82–91
Gouy M (1987) Codon contexts in enterobacterial and coliphage genes. Mol Biol Evol 4:426–444
Grantham R, Gautier C, Gouy M, Mercier R, Pave A (1980) Codon catalog usage and the genome hypothesis. Nucleic Acids Res 8:r49–r62
Gupta SK, Bhattacharyya TK, Ghosh TC (2004) Synonymous codon usage in Lactococcus lactis: mutational bias versus translational selection. J Biomol Struct Dynam 21:527–536
Hatfull GF, Pedulla ML, Jacobs-Sera D et al (2006) Exploring the mycobacteriophage metaproteome: phage genomics as an educational platform. PLoS Genet 2(6):e92, (online, doi: 101371/journalpgen0020092)
Hendrix R (1999) The long evolutionary reach of viruses. Curr Biol 9:9914–9917
Hendrix RW (2003) Bacteriophage genomics. Curr Opin Microbiol 6:506–511
Hendrix R, Smith M, Burns R, Ford M, Hatfull G (1999) Evolutionary relationships among diverse bacteriophages and prophages: all the world’s a phage. Proc Natl Acad Sci USA 96:2192–2197
Ikemura T (1985) Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol 2:13–34
Kliman RM, Hey J (1994) The effects of mutation and natural selection on codon bias in the genes of Drosophila. Genetics 137:1049–1056
Kunisawa T, Kanaya S, Kutter E (1998) Comparison of synonymous codon distribution patterns of bacteriophage and host genomes. DNA Res 5:319–326
Lawrence JG, Hatfull GF, Hendrix RW (2002) Imbroglios of viral taxonomy: genetic exchange and failings of phenetic approaches. J Bacteriol 184:4891–4905
Lee S, Kriakov J, Vilcheze C, Dai Z, Hatfull GF, Jacobs WR Jr (2004) Bxz1: a new generalized transducing phage for mycobacteria FEMS. Microbiol Lett 241:271–276
Liu J, Glazko G, Mushegian A (2006) Protein repertoire of double-stranded DNA bacteriophages. Virus Res 117:68–80
Lü H, Zhao W-M, Zheng Y, Wang H, Qi M, Yu X-P (2005) Analysis of synonymous codon usage bias in Chlamydia. Acta Biochim Biophys Sinica 37:1–10
Médigue C, Rouxel T, Vigier P, Hénaut A, Danchin A (1991) Evidence for horizontal gene transfer in Escherichia coli speciation. J Mol Biol 222:851–856
Miller ES, Heidelberg JF, Eisen JA, Nelson WC, Durkin AS, Ciecko A, Feldblyum TV, White O, Paulsen IT, Nierman WC, Lee J, Szczypinski B, Fraser CM (2003) Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage. J Bacteriol 185(17):5220–5233
Nolan JM, Petrov V, Bertrand C, Krisch HM, Karam JD (2006) Genetic diversity among five T4-like bacteriophages. Virol J 3(30) (doi: 101186/1743-422X-3-30)
Pedulla ML, Ford ME, Houtz JM et al (2003) Origins of highly mosaic mycobacteriophage genomes. Cell 113:171–182
Proux C, van Sinderen D, Suarez J, Garcia P, Ladero V, Fitzgerald GF, Desiere F, Brussow H (2002) The dilemma of phage taxonomy illustrated by comparative genomics of Sfi21-like Siphoviridae in lactic acid bacteria. J Bacteriol 184:6026–6036
Rohwer F, Edwards R (2002) The phage proteomic tree: a genome-based taxonomy for phage. J Bacteriol 184(16):4529–4535
Rokyta DR, Burch CL, Caudle SB, Wichman HA (2006) Horizontal gene transfer and the evolution of microvirid coliphage genomes. J Bacteriol 188(3):1134–1142
Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes CA, Hutchison CA, Slocombe PM, Smith M (1977) Nucleotide sequence of bacteriophage ϕX174 DNA. Nature 265:687–695
Sau K, Gupta SK, Sau S, Ghosh TC (2005) Synonymous codon usage bias in 16 Staphylococcus aureus phages: implication in phage therapy. Virus Res 113(2):123–31
Sharp PM (1986) Molecular evolution of bacteriophage: evidence of selection against recognition sites of host restriction enzymes. Mol Biol Evol 3(1):75–83
Sharp PM, Li W-H (1987) The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acid Res 15:1281–1295
Sharp PM, Rogers MS, McConnell DJ (1985) Selection pressures on codon usage in the complete genome of bacteriophage T7. J Mol Evol 21(2):150–160
Sharp PM, Cowe E, Higgins DG, Shields DC, Wolfe KH, Wright F (1988) Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomices pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Res 16:8207–8211
Sharp PM, Bailes E, Grocock RJ, Peden JF, Sockett RE (2005) Variation in the strength of selected codon usage bias among bacteria. Nucleic Acids Res 33:1141–1153
Shields DC, Sharp PM (1987) Synonymous codon usage in Bacillus subtilis reflects both traditional selection and mutational biases. Nucleic Acids Res 15:8023–8040
Stenico M, Loyd AT, Sharp PM (1994) Codon usage in Caenorhabditis elegans: delineation of translational selection and mutational biases. Nucleic Acids Res 22:2437–2446
Summers WC (2001) Bacteriophage therapy. Annu Rev Microbiol 55:437–451
Varenne S, Buc J, Lloubès R, Lazdunski C (1984) Translation is a non-uniform process Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J Mol Biol 180:549–576
Villarreal L (2001) Persisting viruses could play role in driving host evolution. Am Soc Microbiol News 67:501–507
Weinbauer GM (2004) Ecology of procaryotic viruses. FEMS Microbiol Rev 28:127–181
Weinbauer MG, Rassoulzadegan F (2004) Are viruses driving microbial diversification and diversity? Environ Microbiol 6:1–11
Wichman HA, Badgett MR, Scott LA, Boulianne CM, Bull JJ (1999) Different trajectories of parallel evolution during viral adaptation. Science 285(5426):422–424
Wichman HA, Millstein J, Bull JJ (2005) Adaptive molecular evolution for 13,000 phage generations: a possible arms race. Genetics 170(1):19–31
Willenbrock H, Friis C, Friis AS, Ussery DW (2006) An environmental signature for 323 microbial genomes based on codon adaptation indices. Genome Biol 7:R114
Wood TE, Burke JM, Rieseberg LH (2005) Parallel genotypic adaptation: when evolution repeats itself. Genetica 123:157–170
Acknowledgments
The authors thank Julie Baussand for remarks and help with R and Hervé Isambert for discussions.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Carbone, A. Codon Bias is a Major Factor Explaining Phage Evolution in Translationally Biased Hosts. J Mol Evol 66, 210–223 (2008). https://doi.org/10.1007/s00239-008-9068-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-008-9068-6