Abstract
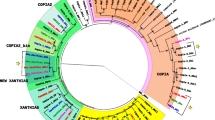
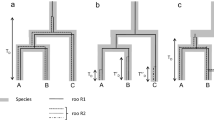
Tirant, a LTR retrotransposon with copies scattered over the chromosome arms of Drosophila melanogaster, is in the process of being lost from the chromosome arms of most natural populations of the sister species D. simulans. In an attempt to clarify the dynamics and evolution of tirant, we have studied the regulatory and reverse transcriptase regions in copies of the nine closely related species of the D. melanogaster subgroup. We show that tirant is mainly vertically transmitted in these species, with the exception of a horizontal transfer event from an ancestor of D. melanogaster to D. teissieri. We propose that, in four of the species (D. melanogaster, D. simulans, D. sechellia, and D. mauritiana), the observed patterns of evolution of the regulatory region vary with genome constraints and with the history and biogeography of the species.






Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Benson G (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580
Biémont C, Cizeron G (1999) Distribution of transposable elements in Drosophila species. Genetica 105:43–62
Biémont C, Vieira C (2005) What transposable elements tell us about genome organization and evolution: The case of Drosophila? Cytogenet Genome Res 110:25–34
Biémont C, Vieira C (2006) Junk DNA as an evolutionary force. Nature 443:521–524
Biémont C, Nardon C, Deceliere G, Lepetit D, Loevenbruck C, Vieira C (2003) Worldwide distribution of transposable element copy number in natural populations of Drosophila simulans. Evolution 57:159–167
Boulesteix M, Weiss M, Biémont C (2006) Differences in genome size between closely related species: the Drosophila melanogaster species subgroup. Mol Biol Evol 23:162–167
Cariou ML (1987) Biochemical phylogeny of the eight species in the Drosophila melanogaster subgroup, including D. sechellia and D. orena. Genet Res 50:181–185
Cizeron G, Lemeunier F, Loevenbruck C, Brehm A, Biémont C (1998) Distribution of the retrotransposable element 412 in Drosophila species. Mol Biol Evol 15:1589–1599
Deceliere G, Charles S, Biémont C (2005) The dynamics of transposable elements in structured populations. Genetics 169:467–474
Di Franco C, Pisano C, Dimitri P, Gigliotti S, Junakovic N (1989) Genomic distribution of copia-like transposable elements in somatic tissue and during development of Drosophila melanogaster. Chromosoma 98:402–410
Fablet M, McDonald JF, Biémont C, Vieira C (2006) Ongoing loss of the tirant transposable element in natural populations of Drosophila simulans. Gene 375:54–62
Galtier N, Gouy M, Gautier C (1996) SeaView and Phylo_Win, two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci 12:543–548
Gonçalves I, Robinson M, Perrière G, Mouchiroud D (1999) JaDis: computing distances between nucleic acid sequences. Bioinformatics 15:424–425
Heredia F, Loreto EL, Valente VL (2004) Complex evolution of gypsy in Drosophilid species. Mol Biol Evol 20:1831–1842
Jordan IK, McDonald JF (1998) Evolution of the copia retrotransposon in the Drosophila melanogaster species subgroup. Mol Biol Evol 15:1160–1171
Kent WJ (2002) BLAT—the BLAST-like alignment tool. Genome Res 12:656–664
Kliman RM, Andolfatto P, Coyne JA, Depaulis F, Kreitman M, Berry AJ, McCarter J, Wakeley J, Hey J (2000) The population genetics of the origin and divergence of the Drosophila simulans complex species. Genetics 156:1913–1931
Kosakovsky Pond SL, Frost SDW, Muse SV (2005) HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679
Lachaise D, Silvain JF (2004) How two Afrotropical endemics made two cosmopolitan human commensals: the Drosophila melanogaster–D. simulans palaeogeographic riddle. Genetica 120:17–39
Lerat E, Rizzon C, Biémont C (2003) Sequence divergence within transposable element families in the Drosophila melanogaster genome. Genome Res 13:1889–1896
Lynch M, Conery JS (2003) The origins of genome complexity. Science 302:1401–1404
Marsano RM, Moschetti R, Caggese C, Lanave C, Barsanti P, Caizzi R (2000) The complete tirant transposable element in Drosophila melanogaster shows a structural relationship with retrovirus-like retrotransposons. Gene 247:87–95
Maruyama K, Hartl DL (1991) Evolution of the transposable element mariner in Drosophila species. Genetics 128:319–329
McClure MA, Johnson MS, Feng DF, Doolittle RF (1988) Sequence comparison of retroviral proteins: relative rates of change and general phylogeny. Proc Natl Acad Sci USA 85:2469–2473
Medstrand P, van de Lagemaat LN, Dunn CA, Landry JR, Svenback D, Mager DL (2005) Impact of transposable elements on the evolution of mammalian gene regulation. Cytogenet. Genome Res 110:342–352
Moltó MD, Paricio N, López-Preciado MA, Semeshin VF, Martínez-Sebastián MJ (1996) Tirant: a new retrotransposon-like element in Drosophila melanogaster. J Mol Evol 42:369–375
Montchamp-Moreau C, Ronsseray S, Jacques M, Lehmann M, Anxolabéhère D (1993) Distribution and conservation of sequences homologous to the 1731 retrotransposon in Drosophila. Mol Biol Evol 10:791–803
Song SU, Gerasimova T, Kurkulos M, Boeke JD, Corces VG (1994) An env-like protein encoded by a Drosophila retroelement: evidence that gypsy is an infectious retrovirus. Genes Dev 8:2046–2057
Strachan T, Coen E, Webb D, Dover G (1982) Modes and rates of change of complex DNA families of Drosophila. J Mol Biol 158:37–54
Tatusova TA, Madden TL (1999) Blast 2 sequences - a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett 174:247–250
Terzian C, Ferraz C, Demaille J, Bucheton A (2000) Evolution of the gypsy endogenous retrovirus in the Drosophila melanogaster subgroup. Mol Biol Evol 17:908–914
Thioulouse J, Chessel D, Dolédec S, Olivier JM (1997) ADE-4: a multivariate analysis and graphical display software. Stat Comput 7:75–83
Uzun O, Gabriel A (2001) A Ty1 transcriptase active-site aspartate mutation blocks transposition but not polymerization. J Virol 75:6337–6347
Vieira C, Biémont C (2004) Transposable element dynamics in two sibling species: Drosophila melanogaster and Drosophila simulans. Genetica 120:115–123
Acknowledgments
We thank Emmanuelle Lerat for useful comments, Gabriel Marais and Jean Thioulouse for their help with the purifying selection tests and the distance matrices, respectively, and Monika Ghosh for reviewing the English text. This work was funded by the Centre National de la Recherche Scientifique (UMR 5558 and GDR 2157 on Transposable Elements).
Author information
Authors and Affiliations
Corresponding author
Additional information
[Reviewing Editor: Dr. Gail Simmons]
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Fablet, M., Souames, S., Biémont, C. et al. Evolutionary Pathways of the tirant LTR Retrotransposon in the Drosophila melanogaster Subgroup of Species. J Mol Evol 64, 438–447 (2007). https://doi.org/10.1007/s00239-006-0108-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-006-0108-9