Abstract
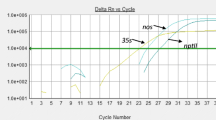
p35S promoter and tNOS terminator are the two primary targets for genetically modified organism (GMO) screening. An increasing number of genetic constructions do not contain p35S and tNOS elements; therefore, new screening assays are required. The use of a larger number of screening methods provides a better coverage of the EU-unapproved GMOs and is a cost-effective approach due to the decrease of tests required for identification. In the present study, new real-time PCR screening assays were developed targeting 10 promoter and terminator elements used in genetically modified constructs: pFMV, pNOS, pSSuAra, pTa29, pUbi, pRice actin, t35S, tE9, tOCS, and tg7. Specificity was verified against different plant species, and the limit of detection was determined on plasmid and genomic reference materials. Criteria of performance were successfully tested taking into account the recommendations of international guidelines. It means that these assays can be considered as ready for an inter-laboratory validation.
Similar content being viewed by others
References
Debode F, Janssen E, Berben G (2007) Physical degradation of genomic DNA of soybean flours does not impair relative quantification of its transgenic content. Eur Food Res Technol 226:273–280
Chaouachi M, Fortabat M-N, Geldreich A, Yot P, Kerlan C, Kebdani N, Audeon C, Romaniuk M, Bertheau Y (2008) An accurate real-time PCR test for the detection and quantification of cauliflower mosaic virus (CaMV) applicable in GMO screening. Eur Food Res Technol 227:789–798
Cankar K, Chauvensy-Ancel V, Fortabat MN, Gruden K, Kobilinsky A, Zel J, Bertheau Y (2008) Detection of nonauthorized genetically modified organisms using differential quantitative polymerase chain reaction: application to 35S in maize. Anal Biochem 376:189–199
Mattarucchi E, Weighardt F, Barbati C, Querci M, Van den Eede G (2005) Development and applications of real-time PCR standards for GMO quantification based on tandem-marker plasmids. Eur Food Res Technol 221:511–519
Wolf C, Scherzinger M, Wurz A, Pauli U, Hübner P, Lüthy J (2000) Detection of cauliflower mosaic virus by the polymerase chain reaction : testing of food components for false-positive 35S-promoter screening results. Eur Food Res Technol 210:367–372
Matsuoka T, Kuribara H, Takubo K, Akiyama H, Miura H, Goda Y, Kusakabe Y, Isshiki K, Toyoda M, Hino A (2002) Detection of recombinant DNA segments introduced to genetically modified maize (Zea mays). J Agric Food Chem 50:2100–2109
Vollenhofer S, Burg K, Schmidt J, Kroath H (1999) Genetically modified organisms in food—screening and specific detection by polymerase chain reaction. J Agric Food Chem 47:5038–5043
Höhne M, Rosa Santisi C, Meyer R (2002) Real-time multiplex PCR: an accurate method for the detection and quantification of 35S-CaMV promoter in genetically modified maize-containing food. Eur Food Res Technol 215:59–64
Zeitler R, Pietsch K, Waiblinger HU (2002) Validation of real-time PCR methods for the quantification of transgenic contaminations in rape seed. Eur Food Res Technol 214:346–351
Alary R, Serin A, Maury D, Jouira HB, Sirven JP, Gautier MF, Joudira P (2002) Comparison of simplex and duplex real-time PCR for quantification of GMO in maize and soybean. Food Control 13:235–244
Kuribara H, Shindo Y, Matsuoka T, Takubo K, Futo S, Aoki N, Hirao T, Akiyama H, Goda Y, Toyoda M, Hino A (2002) Novel reference molecules for quantitation of genetically modified maize and soybean. J AOAC Int 85:1077–1089
Kimio M, Rie A, Naoki S, Masaki S, Hisatsugu I, Kazue S, Takashi T, Kunihiro K, Akihiro H, Kazuo S (2005) Detection of Genetically Modified Organisms in Foreign-made Processed Foods Containing Corn and Potato. J Food Hyg Soc Japan 46:79–85
Fernandez S, Charles-Delobel C, Geldreich A, Berthier G, Boyer F, Collonnier C, Coué-Philippe G, Diolez A, Duplan M-N, Kebdani N, Romaniuk M, Feinberg M, Bertheau Y (2005) Quantification of the 35S Promoter in DNA extracts from genetically modified organisms using real-time polymerase chain reaction and specificity assessment on various genetically modified organisms, part I: operating procedure. J AOAC Int 88:547–573
Reiting R, Broll H, Waiblinger HU, Grohmann L (2007) Collaborative study of a T-nos real-time PCR method for screening of genetically modified organisms in food products. J Verbr Lebensm 2:116–121
Fukuta S, Mizukami Y, Ishida A, Ueda J, Hasegawa M, Hayashi I, Hashimoto M, Kanbe M (2004) Real-time loop-mediated isothermal amplification for the CaMV-35S promoter as a screening method for genetically modified organisms. Eur Food Res Technol 218:496–500
Leimanis S, Hernandez M, Fernandez S, Boyer F, Burns M, Bruderer S, Glouden T, Harris N, Kaeppeli O, Philipp P, Pla M, Puigdomenech P, Vaitilingom M, Bertheau Y, Remacle J (2006) A microarray-based detection system for genetically modified (GM) food ingredients. Plant Mol Biol 61:123–139
Minunni M, Tombelli S, Mariotti E, Mascini M (2001) Biosensors as new analytical tool for detection of Genetically Modified organisms (GMOs). Fresenius Anal Chem 369:589–593
Guo J, Yang L, Chen L, Morisset D, Li X, Pan L, Zhang D (2011) MPIC: a high-throughput analytical method for multiple DNA targets. Anal Chem 83:1579–1586
Babekova R, Funk T, Pecoraro S, Engel K-H, Baikova D, Busch U (2008) Duplex polymerase chain reaction (PCR) for the simultaneous detection of cryIA(b) and the maize ubiquitin promoter in the transgenic rice line KMD1. Biotechnol Biotechnol EQ 22(2008/2):705–708
Pansiot J, Chaouachi M, Cavellini L, Romaniuk M, Ayadi M, Bertheau Y, Laval V (2011) Development of two screening duplex PCR assays for genetically modified organism quantification using multiplex real-time PCR master mixes. Eur Food Res Technol 232:327–334
Dörries H–H, Remus I, Grönewald A, Grönewald C, Harzman C, Berghof-Jäger K (2010) Development of a qualitative, multiplex real-time PCR kit for screening of genetically modified organisms (GMOs). Anal Bioanal Chem 396:2043–2054
ISO 21571:2005. (2005) Foodstuffs. Methods of analysis for the detection of genetically modified organisms and derived products. Nucleic acid extraction. International Organization for Standardization, Geneva
AFNOR XP V03-020-2 (2003) Produits alimentaires. Détection et quantification des organismes végétaux génétiquement modifies et produits dérivés. Partie 2: méthodes basées sur la réaction de polymérisation en chaîne. Norme expérimentale
Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Nuclear Plant Mol Bio Rep 9:208–218
Bennett MD, Leitch IJ (2005) Nuclear DNA amounts in angiosperms: progress, problems and prospects. Ann Bot 95:45–90
Dolezel J, Greilhuber J (2010) Nuclear genome size: are we getting closer? Cytometry A 77:635–642
Praca-Fontes MM, Carvalho CR, Clarindo WR, Cruz CD (2011) Revisiting the DNA C-values of the genome size-standards used in plant flow cytometry to choose the “best primary standards”. Plant Cell Rep 30:1183–1191
GMOSeek project. http://www.nib.si/eng/index.php/aktualno/project/272-gmoseek-better-control-of-genetically-modified-organisms-gmos-for-an-improved-european-food-safety.html. Last consultation on the 5 of March 2012
Block A and Debode F, Grohmann L, Hulin J, Taverniers I, Kluga L, Barbau-Piednoir E, Broeders S, Huber I, Van den Bulcke M, Heinze P, Berben G, Busch U, Roosens N, Janssen E, Žel J, Gruden K, Morisset D (2013) The GMOseek matrix: a decision support tool for optimizing the detection of genetically modified plants (submitted for publication)
Holst-Jensen A, Bertheau Y, De Loose M, Grohmann L, Hamels S, Hougs L, Morisset D, Pecoraro S, Pla M, Van den Bulcke M, Wulff D (2012) Detecting un-authorized genetically modified organisms (GMOs) and derived materials. Biotechnol Adv 30:1318–1335
Querci M, Van den Bulcke M, Žel J, Van den Eede G, Broll H (2008) New approaches in GMO detection. Anal Bioanal Chem 396:1991–2002
Novak PK, Gruden K, Morisset D, Lavrac N, Stebih D, Rotter A, Žel J (2009) GMOtrack: generator of cost-effective GMO testing strategies. J AOAC Int 92:1739–1746
Gerdes L, Busch U, Pecoraro S (2012) GMOfinder—a GMO screening database. Food Anal Meth 5:1368–1376
Spalinskas R, Van den Bulcke M, Van den Eede G, Milcamps A (2012) LT-RADE: an efficient user-friendly genome walking method applied to the molecular characterization of the insertion site of genetically modified maize MON810 and rice LLRICE62. Food Anal Meth. doi:10.1007/s12161-012-9438-y
Morisset D, Broeders S, Block A, Berben G, Huber I, Debode F, Kluga L, Taverniers I, Grohmann L. (2011) Validation guidelines for qualitative methods. Deliverable D8/01. GMOseek project (FSA G03032, SAFEFOODERA)
Cankar K, Ravnikar M, Zel J, Gruden K, Toplak N (2005) Real-time polymerase chain reaction detection of cauliflower mosaic virus to complement the 35S screening assay for genetically modified organisms. J AOAC Int 88:814–822
European Network of GMO Laboratories (2008) Definition of minimum performance requirement for Analytical Methods for GMO testing. CRL GM Food and Feed. http://gmo-crl.jrc.ec.europa.eu/doc/Min_Perf_Requirements_Analytical_methods.pdf. Last consultation on the 5 of March 2012
Guidelines on performance criteria and validation of methods for detection, identification and quantification of specific DNA sequences and specific proteins in food (2010) ALINORM 10/33/23. Appendix III. http://www.codexalimentarius.net/download/report/738/al33_23e.pdf. Last consultation on the 5 of March 2012
Waiblinger H-U, Grohmann L, Mankertz J, Engelbert D, Pietsch K (2010) A practical approach to screen for authorised and unauthorised genetically modified plants. Anal Bioanal Chem 396:2065–2072
Grohmann L, Brunen-Nieweler C, Nemeth A, Waiblinger HU (2009) Collaborative trial validation studies of real-time PCR-Based GMO screening methods for detection of the bar gene and the ctp2-cp4epsps construct. J Agric Food Chem 57:8913–8920
Zhang Y, Zhang D, Li W, Chen J, Peng Y, Cao W (2003) A novel real-time quantitative PCR method using attached universal template probe. Nucleic Acids Res 31(20):e123. doi:10.1093/nar/gng123
Weighardt F, Barbati C, Paoletti C, Querci M, Kay S, De Beuckeleer M, Van den Eede G (2004) Real-time polymerase chain reaction-based approach for quantification of the pat gene in the T25 Zea mays event. J AOAC Int 87:1342–1355
Vaïtilingom M, Pijnenburg H, Gendre F, Brignon P (1999) Real-time quantitative PCR detection of genetically modified Maximizer maize and Roundup Ready soybean in some representative foods. J Agric Food Chem 47:5261–5266
Rho JK, Lee T, Jung SI, Kim TS, Park YH, Kim YM (2004) Qualitative and quantitative PCR methods for detection of three lines of genetically modified potatoes. J Agric Food Chem 52:3269–3274
Randhawa GJ, Chhabra R, Singh M (2010) Decaplex and real-time PCR Based Detection of MON531 and MON15985 Bt cotton events. J Agric Food Chem 58:9875–9881
Dinon AZ, Prins TW, VanDijk JP, Arisi ACM, Scholtens IMJ, Kok EJ (2011) Development and validation of realtime PCR screening methods for detection of cry1A.105 and cry2Ab2 genes in genetically modified organisms. Anal Bioanal Chemi 400:1433–1442
Guan Q, Wang X, Teng D, Yang Y, Tian F, Yin Q, Wang J (2011) Construction of a Standard Reference Plasmid for Detecting GM Cottonseed Meal. Appl Biochem Biotechnol 165:24–34
Acknowledgments
This research was conducted within a Belgian research project (Convention RT-06/6 GMODetec) financed by the Belgian Federal Public Service for Public Health, Food Chain Safety, and Environment in an endeavor involving three partners (ISP/WIV, ILVO, and CRA-W). Isabel Taverniers from ILVO (Belgium) kindly provided Arabidopsis DNA, and DNA from A. tumefaciens strains was kindly provided by Enrico Biondi from the University of Bologna (Italy). A part of this study (pUbi and tE9 targets) was financially supported by the UK Food Standard Agency (FSA, contract G03032), and the German Federal Office of Consumer Protection and Food Safety (BVL) through the project GMOseek, under the European ERA-NET consortium SAFEFOODERA. We are grateful to Marjorie Servais and Laurent Timmermans (HELHA Fleurus) for their technical help. We also thank Denis Roulez, Esther Arranz, Nicaise Kayoka-Mukendi, Cécile Ancion, and Gaëlle Antoine from our technical team (GMO team of CRA-W).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Debode, F., Janssen, E. & Berben, G. Development of 10 new screening PCR assays for GMO detection targeting promoters (pFMV, pNOS, pSSuAra, pTA29, pUbi, pRice actin) and terminators (t35S, tE9, tOCS, tg7). Eur Food Res Technol 236, 659–669 (2013). https://doi.org/10.1007/s00217-013-1921-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-013-1921-1