Abstract
An automated quantum mechanical total line shape (QMTLS) fitting model was implemented for quantitative nuclear magnetic resonance (NMR)-based profiling of 42 metabolites in ultrafiltrated human serum samples. Each metabolite was described by a set of chemical shifts, J-couplings, and line widths. These parameters were optimized for each metabolite in each sample by iteratively minimizing the difference between the calculated and the experimental spectrum. In total, 92.0 to 98.1 % of the signal intensities in the experimental spectrum could be explained by the calculated spectrum. The model was validated by comparison to signal integration of metabolites with isolated signals and by means of standard additions. Metabolites present at average concentration higher than 50 μM were quantified with average absolute relative error less than 10 % when using different initial parameters for the fitting procedure. Furthermore, the biological applicability of the QMTLS model was demonstrated on 287 samples from an intervention study in 37 human volunteers undergoing an exercise challenge. Our automated QMTLS model was able to cope with the large dynamic range of metabolite concentrations in serum and proved to be suitable for high-throughput analysis.

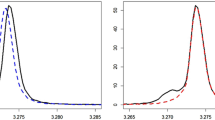
An example of deconvolution with doublets of valine, isoleucine, and keto-leucine and triplets ofleucine and isoleucine a single UF serum sample






Similar content being viewed by others
Abbreviations
- 1H NMR:
-
Proton nuclear magnetic resonance
- QMTLS:
-
Quantum mechanical total line shape fitting
- UF:
-
Ultrafiltration
References
Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, Mandal R, Sinelnikov I, Xia JG, Jia L, Cruz JA, Lim E, Sobsey CA, Shrivastava S, Huang P, Liu P, Fang L, Peng J, Fradette R, Cheng D, Tzur D, Clements M, Lewis A, De Souza A, Zuniga A, Dawe M, Xiong YP, Clive D, Greiner R, Nazyrova A, Shaykhutdinov R, Li L, Vogel HJ, Forsythe I (2009) HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res 37:D603–D610
Zhang AH, Sun H, Wang P, Han Y, Wang XJ (2012) Modern analytical techniques in metabolomics analysis. Analyst 137(2):293–300
Psychogios N, Hau DD, Peng J, Guo AC, Mandal R, Bouatra S, Sinelnikov I, Krishnamurthy R, Eisner R, Gautam B, Young N, Xia JG, Knox C, Dong E, Huang P, Hollander Z, Pedersen TL, Smith SR, Bamforth F, Greiner R, McManus B, Newman JW, Goodfriend T, Wishart DS (2011) The human serum metabolome. PLoS One 6(2):1–23
Barding GA, Salditos R, Larive CK (2012) Quantitative NMR for bioanalysis and metabolomics. Anal Bioanal Chem 404(4):1165–1179
van Duynhoven J, van Velzen E, Jacobs D(2013) In: Graham AW (ed) Annual reports on NMR spectroscopy, vol 80. Elsevier Science
Pauli GF, Godecke T, Jaki BU, Lankin DC (2012) Quantitative H-1 NMR. Development and potential of an analytical method: an update. J Nat Prod 75(4):834–851
Wishart DS (2008) Quantitative metabolomics using NMR. Trac-Trends Anal Chem 27(3):228–237
Zhang SC, Gowda GAN, Ye T, Raftery D (2010) Advances in NMR-based biofluid analysis and metabolite profiling. Analyst 135(7):1490–1498
Tiziani S, Einwas AH, Lodi A, Ludwig C, Bunce CM, Viant MR, Gunther UL (2008) Optimized metabolite extraction from blood serum for H-1 nuclear magnetic resonance spectroscopy. Anal Biochem 377(1):16–23
Stolzenburg S, Lauridsen M, Toft H, Zalloua P, Baunsgaard D (2011) Improved quality of 1H NMR spectroscopic data for enhanced metabolic profiling of low molecular weight metabolites in human serum. Metabolomics 7(2):270–277
Rubtsov DV, Waterman C, Currie RA, Waterfield C, Salazar JD, Wright J, Griffin JL (2010) Application of a Bayesian deconvolution approach for high-resolution H-1 NMR spectra to assessing the metabolic effects of acute phenobarbital exposure in liver tissue. Anal Chem 82(11):4479–4485
Chylla RA, Hu KF, Effinger JJ, Markley JL (2011) Deconvolution of two-dimensional NMR spectra by fast maximum likelihood reconstruction: application to quantitative metabolomics. Anal Chem 83(12):4871–4880
Weljie AM, Newton J, Mercier P, Carlson E, Slupsky CM (2006) Targeted profiling: quantitative analysis of H-1 NMR metabolomics data. Anal Chem 78(13):4430–4442
Saude EJ, Slupsky CM, Sykes BD (2006) Optimization of NMR analysis of biological fluids for quantitative accuracy. Metabolomics 2(3):113–123
Schicho R, Nazyrova A, Shaykhutdinov R, Duggan G, Vogel HJ, Storr M (2010) Quantitative metabolomic profiling of serum and urine in DSS-induced ulcerative colitis of mice by H-1 NMR spectroscopy. J Proteome Res 9(12):6265–6273
Tredwell GD, Behrends V, Geier FM, Liebeke M, Bundy JG (2011) Between-person comparison of metabolite fitting for NMR-based quantitative metabolomics. Anal Chem 83(22):8683–8687
Mercier P, Lewis MJ, Chang D, Baker D, Wishart DS (2011) Towards automatic metabolomic profiling of high-resolution one-dimensional proton NMR spectra. J Biomol NMR 49(3–4):307–323
Schleif FM, Riemer T, Borner U, Schnapka-Hille L, Cross M (2011) Genetic algorithm for shift-uncertainty correction in 1-D NMR-based metabolite identifications and quantifications. Bioinformatics 27(4):524–533
Zheng C, Zhang SC, Ragg S, Raftery D, Vitek O (2011) Identification and quantification of metabolites in H-1 NMR spectra by Bayesian model selection. Bioinformatics 27(12):1637–1644
Aranibar N, Borys M, Mackin NA, Ly V, Abu-Absi N, Abu-Absi S, Niemitz M, Schilling B, Li ZJ, Brock B, Russell RJ, Tymiak A, Reily MD (2011) NMR-based metabolomics of mammalian cell and tissue cultures. J Biomol NMR 49(3–4):195–206
Jukarainen NM, Korhonen SP, Laakso MP, Korolainen MA, Niemitz M, Soininen PP, Tuppurainen K, Vepsalainen J, Pirttila T, Laatikainen R (2008) Quantification of H-1 NMR spectra of human cerebrospinal fluid: a protocol based on constrained total-line-shape analysis. Metabolomics 4(2):150–160
Mierisova S, Ala-Korpela M (2001) MR spectroscopy quantitation: a review of frequency domain methods. NMR Biomed 14(4):247–259
Soininen P, Haarala J, Vepsalainen J, Niemitz M, Laatikainen R (2005) Strategies for organic impurity quantification by H-1 NMR spectroscopy: constrained total-line-shape fitting. Anal Chim Acta 542(2):178–185
Kettunen J, Tukiainen T, Sarin AP, Ortega-Alonso A, Tikkanen E, Lyytikainen LP, Kangas AJ, Soininen P, Wurtz P, Silander K, Dick DM, Rose RJ, Savolainen MJ, Viikari J, Kahonen M, Lehtimaki T, Pietilainen KH, Inouye M, McCarthy MI, Jula A, Eriksson J, Raitakari OT, Salomaa V, Kaprio J, Jarvelin MR, Peltonen L, Perola M, Freimer NB, Ala-Korpela M, Palotie A, Ripatti S (2012) Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nature Genet 44(3):269–276
Soininen P, Kangas AJ, Wurtz P, Tukiainen T, Tynkkynen T, Laatikainen R, Jarvelin MR, Kahonen M, Lehtimaki T, Viikari J, Raitakari OT, Savolainen MJ, Ala-Korpela M (2009) High-throughput serum NMR metabonomics for cost-effective holistic studies on systemic metabolism. Analyst 134(9):1781–1785
Randell RK, Hodgson AB, Lotito SB, Jacobs DM, Boon N, Mela DJ, Jeukendrup AE (2013) No effect of 1 or 7 d of green tea extract ingestion on fat oxidation during exercise. Med Sci Sports Exerc 45(5):883–891, 810.1249/MSS.1240b1013e31827dd31829d31824
Pechlivanis A, Kostidis S, Saraslanidis P, Petridou A, Tsalis G, Veselkov K, Mikros E, Mougios V, Theodoridis GA (2012) 1H NMR study on the short- and long-term impact of two training programs of sprint running on the metabolic fingerprint of human serum. J Proteome Res 12(1):470–480
Tsiafoulis C, Exarchou V, Tziova P, Bairaktari E, Gerothanassis I, Troganis A (2011) A new method for the determination of free l-carnitine in serum samples based on high field single quantum coherence filtering 1H NMR spectroscopy. Anal Bioanal Chem 399(6):2285–2294
Acknowledgments
This research was granted by The Netherlands Metabolomics Centre and The Centre for BioSystems Genomics, both of which are part of The Netherlands Genomics Initiative/Netherlands Organization for Scientific Research. We thank Dr. Rebecca Randell and Dr. Adrian Hodgson from the University of Birmingham for providing the serum samples.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 131 kb)
Rights and permissions
About this article
Cite this article
Mihaleva, V.V., Korhonen, SP., van Duynhoven, J. et al. Automated quantum mechanical total line shape fitting model for quantitative NMR-based profiling of human serum metabolites. Anal Bioanal Chem 406, 3091–3102 (2014). https://doi.org/10.1007/s00216-014-7752-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-014-7752-5