Abstract
Biofilms are complex aggregates formed by microorganisms such as bacteria, fungi and algae, which grow at the interfaces between water and natural or artificial materials. They are actively involved in processes of sorption and desorption of metal ions in water and reflect the environmental conditions in the recent past. Therefore, biofilms can be used as bioindicators of water quality. The goal of this study was to determine whether the biofilms, developed in different aquatic systems, could be successfully discriminated using data on their elemental compositions. Biofilms were grown on natural or polycarbonate materials in flowing water, standing water and seawater bodies. Using an unsupervised technique such as principal component analysis (PCA) and several supervised methods like classification and regression trees (CART), discriminant partial least squares regression (DPLS) and uninformative variable elimination–DPLS (UVE-DPLS), we could confirm the uniqueness of sea biofilms and make a distinction between flowing water and standing water biofilms. The CART, DPLS and UVE-DPLS discriminant models were validated with an independent test set selected either by the Kennard and Stone method or the duplex algorithm. The best model was obtained from CART with 100% correct classification rate for the test set designed by the Kennard and Stone algorithm. With CART, one variable describing the Mg content in the biofilm water phase was found to be important for the discrimination of flowing water and standing water biofilms.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Water quality assessment requires monitoring of carefully selected parameters. Usually water and sediment samples are collected in the hope that their chemical compositions will help to understand the nature of a given local or global environmental event. The chemical analysis of water indicates the water quality at the time of sampling, while analysis of sediments provides information about long-term environmental changes in aquatic systems. Biofilms reflect the environmental conditions in the recent past [1]. They are complex communities composed of microorganisms, which grow at almost any water–substrate interface. Biofilms can accumulate metal ions and play an important role in the processes of sorption and desorption of chemical elements [2]; therefore, they can be very useful bioindicators of water quality.
The aim of this work is to investigate whether the biofilms grown in different aquatic systems can be discriminated on the basis of their chemical compositions. If the discrimination is possible, the next interesting question will be which measured parameters are responsible for it.
For the purpose of the study, biofilm samples originating from different water bodies such as flowing water, standing water or seawater were collected. The elemental concentrations of biofilms and the water phase extracted at the sampling locations were then analysed using inductively coupled plasma optical emission spectrometry (ICP-OES) and inductively coupled plasma mass spectrometry (ICP-MS), respectively. To obtain useful information on the data collected, several pattern recognition techniques are going to be applied. These are principal component analysis (PCA), classification and regression trees (CART), discriminant partial least squares regression (DPLS) and uninformative variable elimination–DPLS (UVE-DPLS). PCA is an unsupervised pattern recognition method that aims to compress and to visualise the data structure, which allows for an easy interpretation of relationships between samples and the measured parameters. The supervised pattern recognition methods like CART, DPLS and UVE-DPLS aim to develop classification or decision rule(s) using a set of samples with known group origin. Then the classification rule(s) determines the belongingness of unknown samples to the available groups [3]. The application of such approaches in our study will provide a better understanding of the accumulation behaviour of different biofilms.
Experimental
Description of the sampling procedure
The biofilm samples investigated can be divided into two groups: a group of systematically sampled biofilms and a group of uniquely sampled biofilms. The samples of the group of systematically sampled biofilms can be further split into two subgroups, namely, biofilms grown on polycarbonate plates and biofilms developed on natural substrates. A detailed description of the samples collected is presented in Table 1.
The biofilm samples collected in the Saale river, in a pond and in the Leutra river were systematically sampled, i.e. the biofilm and water samples were gathered within a definite period of time. The Saale river was chosen as a body of flowing water. The river flows through highly populated regions, which are heavily industrialised and are subjects of typical geogenic and anthropogenic pollution [4]. The Saale water and biofilm samples were collected within a 2-year period (from September 2003 till October 2005). The sampling point was situated in the village of Kunitz located downstream of Jena (Thuringia, Germany). This place was preferred, because it reflects a characteristic pollution of the nearby town. The Saale river is there about 1.5 m deep and Kunitz is remote from civilisation. These conditions facilitated the sampling campaign.
A small pond of 0.75-m depth was selected as a typical example of a standing water body. It was located in the city of Jena. The sampling campaign duration was the same as for the sampling campaign in the Saale river. At these two locations, the biofilms were artificially grown on polycarbonate plates (10 cm × 10 cm) exposed vertically to the water (in the Saale river, in a streaming direction). The plates were fixed into polypropylene boxes, approximately 10 cm under the water surface and 1 m away from the riverbank. After a definite time of exposure, the biofilm samples were immediately transferred into plastic boxes filled with the river or pond water and transported to the laboratory. The plates were then washed with bidistilled water and the biofilm samples were scraped off the whole surface of the polycarbonate plates using a Teflon spatula. The river and pond water samples were collected every 2 weeks.
The Leutra river, located in the city of Jena, was chosen as the second example of a flowing water body. The stony bed of the river, its small depth and good accessibility facilitated the sampling campaign. The biofilm samples from the Leutra river were scraped off the riverbed stones using a plastic spatula. They were placed into polyethylene bottles and transported to the laboratory. The sampling campaign at the Leutra river was held in autumn 2005 and in spring 2006. Additionally, water samples were collected.
The sampling procedure, for water and uniquely sampled biofilms, was the same as that carried out for the Leutra river. The locations of the sampling sites were selected according to the availability of a suitable sampling device. The samples collected were also placed into polyethylene bottles and transported to the laboratory.
Analytical procedure
The biofilm samples collected were air-dried at 105 °C. Then, the samples containing 10–50 mg biofilm powder were dissolved in 3 ml of 70–72% perchloric acid and were heated for 3 h at 50 °C. The remaining dry matter of each sample was further dissolved in water so that the resulting solution was up to 5 ml. A Fisons Instruments (Beverly, MA, USA) Maxim 112 inductively coupled plasma optical emission spectrometer was used to analyse Al, Ca, Fe, K, Mg, Mn, Na and Sr, while Cd, Co, Cr, Cu, Ni, P, Se and Zn were determined with a PerkinElmer (Wellesley, MA, USA) Elan 6000 inductively coupled plasma mass spectrometer. An external aqueous calibration was adopted for the analysis by ICP-OES, while a standard addition procedure was used for the element analysis by ICP-MS. All contents correspond to the sample dry weight. The trueness of the measurements was tested by analysing a certified reference algae material. The element contents were certified for an aqua regia digestion. Additionally, the element contents were determined after microwave digestion with nitric acid. No differences between these two digestion methods were obtained. All the measurements were done in triplicate and the relative standard deviation of the technique was 10–15% for all the biofilms, indicating good repeatability of the measurements.
Theory
Classification and regression trees
The CART method was proposed by Breiman et al. [5], for data modelling and classification. Depending on the type of the response variable, y (categorical or continuous), either classification or regression trees are built. In the present study, we will focus on constructing classification trees only. The goal of CART is to form a set of mutually exclusive regions in the data space, containing as homogeneous groups of objects as possible. This is achieved by finding optimal splits of some suitable explanatory variables at a given threshold value, such that a defined impurity function is minimised. The impurity function measures the homogeneity of each node obtained from the split. It takes the lowest value for pure nodes [5]. The nodes are split while a specified number of objects are not present in the child nodes or the nodes are not pure. A node which cannot be split any further is called a terminal node. One of the most popular impurity functions is entropy [5] and it is the function used in our study.
Owing to a binary data splitting, the results of CART can easily be visualised as a binary tree, which consists of a number of nodes symbolising subgroups of data objects.
In order to ensure good prediction properties of the constructed tree, the number of the tree nodes should be optimal. Selection of the optimal number of nodes relies upon a deletion of some nodes from the tree, which is done by means of the so-called cost-complexity pruning [5].
Discriminant partial least squares
The DPLS approach aims to relate a set of n explanatory variables (predictors), X (m × n), to a dependent variable, y. The dependent variable, y, is either a discrete variable, representing the belongingness of m objects to two defined groups denoted by −1 and 1, or a binary variable.
The popularity of the DPLS method in chemometrics is due to its attractive properties. DPLS can successfully deal with multicollinearity in the data by constructing a few (f) latent factors, T (m × f), which maximise the covariance between X and y [6].
In order to obtain DPLS models with good prediction abilities, an optimal number of factors should be chosen. The optimal number of factors is usually found with the help of a cross-validation procedure [6]. The model with the smallest root mean square error of cross-validation (RMSCV) is to be selected. The goodness of model fit is indicated by a root mean square (RMS) error, whereas the success of the prediction is expressed by a root mean square error of the test set. Moreover, the performance of DPLS depends on the set of samples used for its construction. The model set has to cover all possible sources of data variance. Furthermore, DPLS is sensitive to the number of objects used to build the model. Its performance is optimal when the model set contains two groups with the same number of objects [7]. The predictive ability of the model built also depends on the quality of the variables measured.
Uninformative variable elimination–discriminant partial least squares
Usually, the samples collected are characterised by a large number of variables in order to ensure a detailed description of the event studied. However, some of the experimental variables may be irrelevant for the particular discriminant problem. Such uninformative variables, which have a high variance, but small covariance with y, lead to a DPLS model with unsatisfactory predictive ability. Therefore, finding an optimal set of variables by discarding the uninformative variables from the data can substantially improve the DPLS model by a decrease of the prediction error for test samples or/and a decrease of model complexity. The variable selection method used in our study is UVE-DPLS [8]. With UVE-DPLS, variables with unstable regression coefficients are removed. In order to estimate the stability of the regression coefficients, a matrix, N (m × p), containing at least p=300 random variables, is augmented with the matrix of experimental variables, X (m × n), which results in a matrix, Z, of dimension m × n+p. To keep the influence of the variables added negligible, their elements are generated from the normal distribution and are multiplied by a small constant with magnitude 1 × 10–10. The matrix of regression coefficients is constructed by the use of a leave-one-out cross-validation procedure, i.e. m PLS models are built and each one by using m-1 objects. The stability of a variable is determined by the ratio of the mean of m regression coefficients and their standard deviation. The variables with absolute stabilities of regression coefficients below a given cutoff value are uninformative and are deleted from the data. The cutoff value is defined as the largest absolute value of all stability values for the random variables added.
The goodness of a discrimination model is characterised by the percentage of correct classification or the so-called correct classification rate. It is commonly agreed that the higher the correct classification rate, the better the model. Additionally, one should consider sensitivity and selectivity of the model. For a two-class problem for instance, sensitivity is defined as the percentage of correctly classified samples of class A, while selectivity is the percentage of correctly classified samples of class B.
Results and discussion
All the data collected were organised in a matrix, X, of dimension 71 × 34. All 71 biofilm samples (Table 1) were characterised by the contents of 17 chemical elements analysed in the biofilms and in the water phases extracted at the sampling locations. Three groups of samples were distinguished depending on the type of water body, namely flowing water, standing water and seawater.
Firstly, PCA was used for an overall exploration of the data structure. PCA is an unsupervised approach, and is frequently employed for data compression and visualisation [9]. With PCA, the original data matrix, X (m × n), is decomposed into two matrices: a scores matrix, T (m × n), the columns of which contain principal components (PCs) and a loadings matrix, P (n × n). PCs are found as linear combinations of explanatory variables by maximising the variance of projected data. The loadings matrix, P, describes the contributions of each variable to the constructed PCs.
Prior to the PCA analysis, the explanatory variables were autoscaled, because they had been measured in different units. Autoscaling is performed by subtracting the column mean from each data element and dividing it by the corresponding standard deviation. It gives variables the same importance in the PCA analysis. The results of PCA for autoscaled data are presented in Fig. 1.
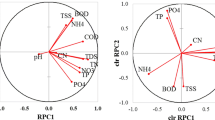
Principal component analysis of the data set containing the uniquely sampled biofilms and the systematically sampled biofilms: a scree plot of the cumulative percentage of data variance explained by the consecutive principal components (PC), b projection of biofilms on the plane defined by PC 1 and PC 2, c projection of biofilms on the plane defined by PC 1 and PC 3, d projection of variables on the plane defined by PC 1 and PC 2 and e projection of variables on the plane defined by PC 1 and PC 3
The first three PCs explain about 50% of the total data variance (Fig. 1a). Figure 1a indicates that the compression is not very effective, because the data variance is distributed over all PCs. However, some general trends in the data structure can be revealed.
All the seawater samples are differentiated from the standing water and flowing water samples in the PC 1–PC 2 score plot (Fig. 1b). The sea samples can be divided into two subgroups. The first subgroup contains the samples from Steinbeck (Germany), Travemünde (Germany) and Damp (Germany), located in the Baltic Sea, while the second subgroup includes the samples from Punta Skala (Croatia), Nin (Croatia) and Majorca (Spain), situated in the Mediterranean Sea. The standing water and flowing water biofilm samples overlap. The flowing water biofilm sample from Munich can also be distinguished from all the other samples along PC 2. This distinction is even more evident along PC 3 (Fig. 1c). Looking at the loading plots, shown in Fig. 1d and e, one finds the reasons for the objects’ distributions observed in the score plots. PC 1 represents the Sr, Cu, Mg, Se, K and Na contents in the water phase (W-Sr, W-Cu, W-Mg, W-Se, W-K, W-Na). This factor can be related to the salt content of the water phase and is conditionally called the ‘salt’ factor. The second PC, PC 2, is mainly associated with Fe and Mg (Fig. 1d). These elements are basic components participating in the biofilm formation. PC 3 reflects the Mn, Zn, Cd, Pb, Fe, and Co contents in the water phase. The presence of Cd and Pb is usually a result of environmental contamination and that is why PC 3 is associated with the anthropogenic influence. From the information obtained from the score and loading plots, it follows that the water phase of the biofilm grown in seawater is indeed richer in dissolved salts than the water phase of the standing water and flowing water bodies. The levels of Fe and Mg in sea biofilms are also higher in comparison with those for the other biofilms. The salt content, pH and temperature of water vary at different sampling locations and they influence the biofilm formation. It was reported in [10] that Mg strongly influences attachment and biofilm structure. The surface colonisation and biofilm depth increase with the increasing Mg concentration. The biofilms collected in Punta Skala, Nin and Majorca contain higher levels of dissolved salts in the water phase and higher Fe and Mg contents in comparison with the biofilms collected in Travemünde, Damp and Steinbeck. The sample originating from Munich shows a high anthropogenic influence, i.e. it has higher Mn, Zn, Cd, Pb, Fe and Co contents in the water phase and lower Fe and Mg contents in the biofilm in comparison with the other samples.
In order to see whether the biofilms developed in standing water could be distinguished from the biofilms grown in flowing water, supervised approaches such as CART, DPLS and UVE-DPLS were applied. Furthermore, it was important to determine if the models constructed could predict the origin of new biofilm samples and how well. Another question to be answered was what variables are responsible for an eventual discrimination of groups. Only seven biofilms were grown in seawater; therefore, they were excluded from the forthcoming analysis.
To construct a reliable discriminant model and to test its predictive ability, the data were divided into two subsets (model and test) with the Kennard and Stone [11, 12] and duplex [13] algorithms enabling a uniform subset selection. In the Kennard and Stone method, objects in the model set are selected sequentially, starting with the object closest to the data mean. The next object included in the subset is the one situated furthest away from the first one. The third object selected is the most distant one from the objects selected in the model set. The selection of objects continues while a predefined number of objects are not assigned to the model set. The remaining objects form the test set. As a similarity measure, the Euclidian distance was used. With the duplex algorithm, the two most distant objects in the data are found and included in the model set. The next two most distant objects are assigned to the test set. The remaining objects are consecutively added to the subsets, switching over to the most distant unassigned object with respect to the model set and to the most distant unassigned object with respect to the test set. The Kennard and Stone algorithm ensures that the objects in the model set cover all possible sources of data variance, while the duplex method guarantees the representativeness of both subsets. Selection of model and test sets should be done for each group separately. When a preprocessing procedure is required, the selection of objects is applied to preprocessed data. In our study, the model and test sets were selected using autoscaled data in order to remove the scale differences among variables while evaluating the Euclidean distances among objects. It should be mentioned that the performance of CART is not influenced by autoscaling. In our study, the model set of dimension 42 × 34 contains 21 flowing water and 21 standing water biofilm samples, whereas the test set of dimension 22 × 34 includes 15 biofilms of flowing water and seven biofilms of standing water.
Results of CART, DPLS and UVE-DPLS for model and test sets designed with the Kennard and Stone algorithm
To trace the importance of variables responsible for the discrimination of both groups, a classification tree was built. After tenfold cross-validation, an optimal tree, containing two terminal nodes, was selected. The cross-validation error is 14%, indicating a relatively good predictive ability of the constructed tree shown in Fig. 2.
Since there is only one split in the tree, the discriminant problem is rather simple and the most discriminative variable describes the Mg content in the water phase (W-Mg). As mentioned before, Mg plays an important role during the biofilm formation [10]. All the model set samples, belonging to the group of standing water (17 samples), have Mg content in water phase below 37 mg g−1. The remaining samples (21 flowing water biofilms and four standing water biofilms) are placed in the left terminal node, which results in a misclassification error of 9.5% for the complete tree. Although four model standing water biofilms are recognised as flowing water biofilms, the constructed classification tree provides a correct classification of 100% for the test samples (Table 2). Therefore, the model yields fairly high sensitivity (percentage of correct classification of the test flowing water biofilms) and selectivity (percentage of correct classification of the test standing water biofilms).
Additionally, good discrimination results can be obtained when the primary split is made on the variable describing the Ca content in the water phase (W-Ca). This variable is a competitive variable selected after removing W-Mg. The split on W-Ca leads to a total misclassification error of 14.3%. The presence of Ca has been shown to have an influence on mechanical properties of biofilms [14].
In the next step of the investigation, DPLS was considered, in order to check if a discrimination model using linear combinations of explanatory variables can perform better than CART. The DPLS model has complexity 1. The RMSCV is 0.95 and RMS error is 0.64. The DPLS model constructed allows for 81.8% correct classification of the test set samples. The analysis of the misclassified test samples indicates that four out of 15 (26.7%) flowing water biofilms collected in the Leutra river are incorrectly predicted as standing water biofilms; therefore, the model has a lower sensitivity (73.3%) than the CART model. All the test samples belonging to the group of standing water biofilms are well predicted, which again indicates the high selectivity (100%) of the model constructed (Table 2). An improved DPLS model was obtained by use of the UVE-DPLS approach, after discarding the uninformative variables. The one-factor UVE-DPLS model constructed with three informative variables (W-Mg, W-Ca, W-Sr), offers a total correct classification of 90.9% for the test set. It yields a selectivity of 100% and a better sensitivity (86.7%) in comparison with the DPLS model, because only two out of 15 (13.3%) biofilms grown in the flowing water of the Leutra river are now assigned to the group of standing water biofilms (Table 2).
The best discrimination results are obtained from CART, even though this model shows a misclassification error of 9.5% for the complete tree. Since the splits are done in a univariate way, the correlation between variables is not taken into account. Therefore, CART provides unsatisfactory results when a linear combination of variables is responsible for discriminating the samples. This, however, cannot be verified unless multivariate approaches such as DPLS and UVE-DPLS are used. Although CART and UVE-DPLS have different objective functions, common variables are selected as essential for the discrimination. The primary variable, W-Mg, and two competitive variables, W-Ca and W-Sr, in CART are also selected by UVE-DPLS.
Results of CART, DPLS and UVE-DPLS for model and test sets designed with the duplex algorithm
Results of CART, DPLS and UVE-DPLS were obtained using data designed with the duplex algorithm, which ensures the representativeness of the model and test sets.
The classification tree built has two terminal nodes and the primary split is again made on the variable representing the Mg content (W-Mg) in the water phase. The cross-validation error is 7.1%. Two out of 42 model set samples are wrongly classified, which leads to a misclassification error of 4.8% for the complete tree. Compared with the previous results, the constructed tree shows a better performance for the model set samples, but worse prediction rates (Table 2); therefore, the model has again a fairly high sensitivity (100%), but quite low selectivity (57.1%).
The DPLS model constructed for the data designed by the duplex algorithm shows slightly better prediction ability (86.4%) than the model built for the data designed by the Kennard and Stone algorithm (81.6%). It presents a better sensitivity (100%), but a reduced selectivity, with only 57.1% of standing water samples being well recognised. A discriminant model characterised by relatively high sensitivity and selectivity parameters is to be preferred over a model with a high sensitivity and a low selectivity. Therefore, the UVE-DPLS model for data designed by the Kennard and Stone algorithm is to be favoured (Table 2). All the methods allow a correct prediction for 86.4% of samples. The samples collected at Chemnitz and White Dak Pond, Metebach, are improperly classified by all methods. In fact, this is not a striking observation though when the data contain some samples that are different in comparison with the majority of samples. These samples are always assigned to the model set using the Kennard and Stone method and then the test samples are correctly predicted. Using the duplex method, we assigned some atypical samples to the test set, which results in a construction of models with too pessimistic predictive abilities.
Results of CART, DPLS and UVE-DPLS for biofilm samples grown on natural substrates
Another important issue to be discussed is whether the biofilm samples grown on natural substrates (see the group of uniquely sampled biofilms in Table 1) can be used to derive similar conclusions as those drawn using the whole data. If this is possible, the sampling procedure will be carried out in a simpler way, which will be less time-consuming and relatively low in price.
For an initial inspection of the data structure, PCA was considered. PCA was applied to autoscaled data (25 × 34) containing only uniquely sampled biofilms and the results are presented in Fig. 3
Principal component analysis of the data set containing the uniquely sampled biofilms: a scree plot of the cumulative percentage of data variance explained by the consecutive PCs, b projection of biofilms on the plane defined by PC 1 and PC 2, c projection of biofilms on the plane defined by PC 1 and PC 3, d projection of variables on the plane defined by PC 1 and PC 2 and e projection of variables on the plane defined by PC 1 and PC 3
The first three PCs account for 59.1% of the total data variance (Fig. 3a). Similar to PCA of the whole data, the compression is not very effective. The biofilms grown in seawater can again be distinguished along PC 1 (Fig. 3b). Moreover, two subgroups of sea biofilms are distinguished along PC 1 (Fig. 3c). The content of the subgroups is the same as before. The biofilm sample collected in Munich is again found far away from all the other samples. Another extreme biofilm sample, collected in the Aller river (Celle, Germany), appears along PC 3. Regarding the variable loadings (Fig. 3d, e), PC 1 is again associated with the salt content of the biofilm water phases, while PC 2 probably is now linked to the contamination of the biofilm water phases, because the variables W-Zn, W-Cd, W-Ni, W-Fe, W-Co, W-Mn and W-Pb possess high loading values. PC 3 consists of Zn, Cd and Pb, which are usually associated with an anthropogenic influence and this factor is therefore associated with contaminants accumulated by the biofilm. Summarising the results of PCA, one can additionally point out that the biofilm samples collected in Majorca, Punta Skala and Nin are richer in Zn, Cd and Pb in comparison with the remaining sea biofilm samples. Moreover, the highest Zn, Cd and Pb contents are characteristic for the biofilm sample from Celle.
In order to construct the CART, DPLS and UVE-DPLS models, only data of natural biofilms grown in flowing water and in standing water were considered. Since the number of samples in each group is small (Table 1), the models were used for an exploratory purpose only. Because of this, the predictive abilities of the models were not tested using an independent test set.
The complete classification tree with three terminal nodes is shown in Fig. 4. The primary split is made on the variable describing the Pb content in the water phase (W-Pb). W-Pb is the most discriminant variable. The next split on variable Al corrects the improper assignment of one sample and it is of a lower importance. Owing to the small number of samples, the required tenfold cross-validation procedure could not be applied and, therefore, the cross-validation error was not reported. All the biofilms grown in standing water are well classified, but two biofilm samples grown in flowing water are wrongly classified, which results in a total classification rate of 88.9%. The incorrectly classified samples originate from Steinach (Germany) and Geithain (Germany).
The DPLS model constructed has complexity 1. RMSCV is 1.78 and RMS error accounts for 0.59. Two samples are incorrectly classified. One of them belongs to the biofilms of standing water and originates from Chemnitz (Germany), while the other one is the biofilm collected in the flowing water body (São Lourenço) located in Juquitiba (Brazil). The DPLS model built yields a total classification rate of 88.9%. It should be emphasised that DPLS can lead to a too optimistic result when the number of variables outnumbers the number of samples [7]. A remedy for this problem is to reduce the number of variables by the use of a feature selection technique, e.g. UVE-DPLS. The UVE-DPLS model has RMSCV of 0.95. One variable, namely W-Pb, is selected. However, all biofilms grown in flowing water are correctly classified with the model constructed, but all biofilms grown in standing water are improperly classified.
Conclusions
Discrimination between sea biofilms and the remaining standing water and flowing water biofilms is straightforward by investigating the score plots obtained from PCA. The loading plots emphasise the expected higher salt content of the water phases extracted from the sea biofilms as well as their higher levels of Fe and Mg in comparison with the other biofilms. A further discrimination between flowing water and standing water biofilms is possible by means of supervised methods like CART, DPLS and UVE-DPLS. The best discriminant model is obtained from CART. One variable describing the Mg content in the water phase (W-Mg) is enough to build a model with 9.5% misclassification error. All test samples selected by the Kennard and Stone algorithm are correctly classified using the constructed CART model. The DPLS and UVE-DPLS methods do not outperform CART for the data set studied and, therefore, it can be pointed out that a linear combination of explanatory variables does not lead to a better prediction for new samples. Moreover, CART appears as a very simple and efficient discriminant technique leading to a straightforward data interpretation in terms of explanatory variables. Hence, CART can be considered as a pilot discriminant approach. When the CART model is not satisfactory, one can apply discriminant methods, such as DPLS and UVE-DPLS, or if necessary to use a nonlinear multivariate classifier like, e.g., support vector machines.
All discriminant models, CART, DPLS and UVE-DPLS, lead to 86.4% correct classification for the test set designed by the duplex algorithm. However, CART uses only one variable (W-Mg), UVE-DPLS selects nine variables and DPLS uses all explanatory variables to build the model.
Discrimination of flowing water and standing water biofilms that are uniquely sampled, using CART, DPLS and UVE-DPLS models, is done only for a better understanding of the data collected. For a definite conclusion whether these two groups of samples can be discriminated, more samples are required to properly validate the discriminant models.
References
Mages M, von Tümpling W, van der Veen A, Baborowski M (2006) Spectrochimica Acta Part B 61:1146–1152
Bryers JD, Characklis W (1990) In: Characklis WG, Marshall KC (eds) Biofilms. Wiley, New York, pp 671–696
Massart DL, Kaufman L (1983) Interpretation of analytical data by the use of cluster analysis. Krieger, Malbar
Einax JW, Truckenbrodt D, Kampe O (1998) Microchem J 58:315–324
Breiman L, Olshen JH, Stone CG (1984) Classification and regression trees. Wadsworth International, Belmont
Martens H, Næs T (1989) Mutivariate calibration. Wiley, Chichester
Brereton R (2006) Trends Aanal Chem 25:1103–1111
Centner V, Massart DL, de Noord O, de Jong S, Vandeginste BM, Stema C (1996) Anal Chem 68:3851–3858
Vandeginste BMG, Massart DL, Buydens LMC, de Jong S, Lewi PJ, Smeyers-Verbeke J (1998) .Handbook of chemometrics and qualimetrics, part B. Elsevier, Amsterdam
Song B, Leff L (2006) Microbiol Res 161:355–361
Kennard RW, Stone LA (1969) Technometrics 11:137–148
Daszykowski M, Walczak B, Massart DL (2002) Anal Chim Acta 468:91–103
Snee RD (1977) Technometrics 19:415–428
Lattner D, Flemming HC, Mayer C (2003) Int J Biol Macromol 33:81–88
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Stanimirova, I., Kubik, A., Walczak, B. et al. Discrimination of biofilm samples using pattern recognition techniques. Anal Bioanal Chem 390, 1273–1282 (2008). https://doi.org/10.1007/s00216-007-1648-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-007-1648-6