Abstract
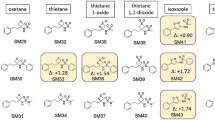
The REGDIA regression diagnostics algorithm in S-Plus is introduced in order to examine the accuracy of pK a predictions made with four updated programs: PALLAS, MARVIN, ACD/pKa and SPARC. This report reviews the current status of computational tools for predicting the pK a values of organic drug-like compounds. Outlier predicted pK a values correspond to molecules that are poorly characterized by the pK a prediction program concerned. The statistical detection of outliers can fail because of masking and swamping effects. The Williams graph was selected to give the most reliable detection of outliers. Six statistical characteristics (F exp, R 2, \( {\text{R}}^{2}_{{\text{P}}} \), MEP, AIC, and s(e) in pK a units) of the results obtained when four selected pK a prediction algorithms were applied to three datasets were examined. The highest values of F exp, R 2, \( {\text{R}}^{2}_{{\text{P}}} \), the lowest values of MEP and s(e), and the most negative AIC were found using the ACD/pK a algorithm for pK a prediction, so this algorithm achieves the best predictive power and the most accurate results. The proposed accuracy test performed by the REGDIA program can also be applied to test the accuracy of other predicted values, such as log P, log D, aqueous solubility or certain physicochemical properties of drug molecules.





Similar content being viewed by others
References
Xing L, Glen RC (2002) Novel methods for the prediction of logP, pK and logD. J Chem Inf Comput Sci 42:796–805
Xing L, Glen RC, Clark RD (2003) Predicting pKa by molecular tree structured fingerprints and PLS. J Chem Inf Comput Sci 43:870–879
Tajkhorshid E, Paizs B, Suhai (1999) Role of isomerization barriers in the pK a control of the retinal schiff base: a density functional study. J Phys Chem B 103:4518–4527
Tripos (2007) SYBYL software. Tripos, Inc., St. Louis, MO (http://www.tripos.com, cited 25 July 2007)
ACD/Labs (2007) pKa Predictor 3.0. Advanced Chemistry Development Inc., Toronto, Canada (http://www.acdlabs.com, cited 25 July 2007)
Rekker RF, ter Laak AM, Mannhold R (1993) Prediction by the ACD/pK a method of values of the acid–base dissociation constant (pK a) for 22 drugs. Quant Struct–Act Relat 12:152
Slater B, McCormack A, Avdeef A, Commer JEA (1994) Comparison of ACD/pKa with experimental values. Pharm Sci 83:1280–1283
ACD/Labs (1997) Results of titrometric measurements on selected drugs compared to ACD/pK a September 1998 predictions (poster). In: AAPS, 1–6 November 1997, Boston, MA
Szegezdi J, Csizmadia F (2004) Marvin plug-in. In: Prediction of dissociation constant using microconstants. http://www.chemaxon.com/conf/Prediction_of_dissociation_constant_using_microconstants.pdf. Cited 25 July 2007
Gulyás Z, Pöcze G, Petz A, Darvas F PALLAS cluster—a new solution to accelerate the high-throughput ADME-TOX prediction. CompuDrug Chemistry Ltd., Sedona, AZ (see http://www.compudrug.com, last cited 25 July 2007)
Kim KH, Martin YC (1991) Direct prediction of linear free energy substituent effects from 3D structures using comparative molecular field effect. 1: Electronic effect of substituted benzoic acids. J Org Chem 56:2723–2729
Kim KH, Martin YC (1991) Direct prediction of dissociation constants of clonidine-like imidazolines, 2-substituted imidazoles, and 1-methyl-2-substituted imidazoles from 3D structures using a comparative molecular field analysis (CoMFA) approach. J Med Chem 34:2056–2060
Gargallo R, Sotriffer CA, Liedl KR, Rode BM (1999) Application of multivariate data analysis methods to comparative molecular field analysis (CoMFA) data: proton affinities and pK a prediction for nucleic acids components. J Comput Aided Mol Des 13:611–623
Perrin DD, Dempsey B, Serjeant EP (1981) pK a prediction for organic acids and bases. Chapman and Hall, London
Habibi-Yangjeh A, Danandeh-Jenagharad M, Nooshyar M (2005) Prediction acidity constant of various benzoic acids and phenols in water using linear and nonlinear QSPR models. Bull Korean Chem Soc 26:2007–2016
Popelier PLA, Smith PJ (2006) QSAR models based on quantum topological molecular similarity. European J Med Chem 41:862–873
Hilal SH, Karickhoff SW, Carreira LA (2003) Prediction of chemical reactivity parameters and physical properties of organic compounds from molecular structure using SPARC (EPA/600/R-03/030 March 2003). National Exposure Research Laboratory, Office of Research and Development, US Environmental Protection Agency, Research Triangle Park, NC
Meloun M, Bordovská S, Kupka K (2007) Outliers detection in the statistical accuracy test of a pK a prediction. Anal Chim Acta (in press)
MathSoft (1997) S-PLUS. MathSoft, Seattle, WA (see http://www.insightful.com/products/splus, cited 25 July 2007)
Meloun M, Militký J, Forina M (1992–1994) Chemometrics for analytical chemistry, vols 1–2. Ellis Horwood, Chichester, UK
ACD/Labs (2007) ACD/pK a DB vs. experiment: a comparison of predicted and experimental values. http://www.acdlabs.com/products/phys_chem_lab/pka/exp.html. Cited 25 July 2007
Lombardo F, Obach RS, Shalaeva MY, Feng G (2004) Prediction of human volume of distribution values for neutral and basic drugs. 2: Extended dataset and leave-class-out statistics. J Med Chem 47:1242–1250
Luan F, Ma W, Zhang H, Zhang X, Liu M, Hu Z, Fan B (2005) Prediction of pK a for neutral and basic drugs based on radial basis function neutral networks and the heuristic method. Pharm Research 22:1454–1460
Masuda T, Jikihara T, Nakamura K, Kimura A, Takagi T, Fujiwara H (1997) Introduction of solvent-accessible surface area in the calculation of the hydrophobicity parameter log P from an atomistic approach. J Pharm Sciences 86:57–63
Moriguchi I, Hirono S, Nakagome I, Hirano H (1994) Comparison of reliability of log P values for drugs calculated by several methods. Chem Pharm Bull 42:976–978
Leo AJ (1995) Critique of recent comparison of log P calculation methods. Chem Pharm Bull 43:512–513
Suzuki T, Kudo Y (1990) Automatic log P estimation based on combined additive modeling methods. J Comput Aided Mol Design 4:155–198
Kolovanov EA, Petrauskas AA (2007) Comparison of the accuracy of log P and log D calculations for 22 drugs. http://www.acdlabs.com/publish/acc_logp.html. Cited 25 July 2007
Kolovanov EA, Petrauskas AA (2007) Re-evaluation of log P data for 22 drugs and comparison of six calculation methods. http://www.acdlabs.com/publish/ac_logp.html. Cited 25 July 2007
Hansen NT, Kouskoumvekaki I, Jorgensen FS, Brunak S, Jonsdottir SO (2006) Prediction of pH-dependent aqueous solubility of druglike molecules. J Chem Inf Model 46:2601–2609
Engkvist O, Wrede P (2002) High-throughput, in silico prediction of aqueous solubility based on one- and two-dimensional descriptors. J Chem Inf Comput Sci 42:1247–1249
Acknowledgements
The financial support of the Czech Ministry of Education (Grant No MSM0021627502) and of the Grant Agency of the Czech Republic (Grant No NR 9055-4/2006) is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Meloun, M., Bordovská, S. Benchmarking and validating algorithms that estimate pK a values of drugs based on their molecular structures. Anal Bioanal Chem 389, 1267–1281 (2007). https://doi.org/10.1007/s00216-007-1502-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-007-1502-x