Abstract
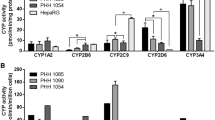
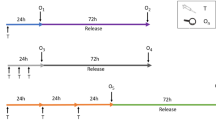
Toxicology is facing a major change in the way toxicity testing is conducted by moving away from animal experimentation towards animal-free methods. To improve the in vitro genotoxicity assessment of chemical and physical compounds, there is an urgent need to accelerate the development of 3D cell models in high-throughput DNA damage detection platforms. Among the alternative methods, hepatic cell lines are a relevant in vitro model for studying the functions of the liver. 3D HepaRG spheroids show improved hepatocyte differentiation, longevity, and functionality compared with 2D HepaRG cultures and are therefore a relevant model for predicting in vivo responses. Recently, the comet assay was developed on 3D HepaRG cells. However, this approach is still low throughput and does not meet the challenge of evaluating the toxicity and risk to humans of tens of thousands of compounds. In this study, we evaluated the performance of the high-throughput in vitro CometChip assay on 2D and 3D HepaRG cells. HepaRG cells were exposed for 48 h to several compounds (methyl methanesulfonate, etoposide, benzo[a]pyrene, cyclophosphamide, 7,12-dimethylbenz[a]anthracene, 2-acetylaminofluorene, and acrylamide) known to have different genotoxic modes of action. The resulting dose responses were quantified using benchmark dose modelling. DNA damage was observed for all compounds except 2-AAF in 2D HepaRG cells and etoposide in 3D HepaRG cells. Results indicate that the platform is capable of reliably identifying genotoxicants in 3D HepaRG cells, and provide further insights regarding specific responses of 2D and 3D models.



Similar content being viewed by others
References
Anthérieu S, Chesné C, Li R et al (2010) Stable expression, activity, and inducibility of cytochromes P450 in differentiated HepaRG cells. Drug Metab Dispos 38:516–525. https://doi.org/10.1124/dmd.109.030197
Ates G, Mertens B, Heymans A et al (2018) A novel genotoxin-specific qPCR array based on the metabolically competent human HepaRG™ cell line as a rapid and reliable tool for improved in vitro hazard assessment. Arch Toxicol 92:1593–1608. https://doi.org/10.1007/s00204-018-2172-5
Baird WM, Hooven LA, Mahadevan B (2005) Carcinogenic polycyclic aromatic hydrocarbon-DNA adducts and mechanism of action. Environ Mol Mutagen 45:106–114. https://doi.org/10.1002/em.20095
Buick JK, Williams A, Gagné R et al (2020) Flow cytometric micronucleus assay and TGx-DDI transcriptomic biomarker analysis of ten genotoxic and non-genotoxic chemicals in human HepaRG™ cells. Genes Environ 42:1–14. https://doi.org/10.1186/s41021-019-0139-2
Buick JK, Williams A, Meier MJ et al (2021) A modern genotoxicity testing paradigm: integration of the high-throughput CometChip® and the TGx-DDI transcriptomic biomarker in human HepaRG™ cell cultures. Front Public Heal 9:1–19. https://doi.org/10.3389/fpubh.2021.694834
Chao C, Engelward BP (2020) Applications of CometChip for environmental health studies. Chem Res Toxicol 33:1528–1538. https://doi.org/10.1021/acs.chemrestox.9b00393
Guillouzo A, Guguen-Guillouzo C (2018) HepaRG cells as a model for hepatotoxicity studies. Stem cells birth defects research development toxicology. Wiley, Oxford, pp 309–339
Gunness P, Mueller D, Shevchenko V et al (2013) 3D organotypic cultures of human heparg cells: a tool for in vitro toxicity studies. Toxicol Sci 133:67–78. https://doi.org/10.1093/toxsci/kft021
Hardy A, Benford D, Halldorsson T et al (2017) Update: use of the benchmark dose approach in risk assessment. EFSA J 15:1–41. https://doi.org/10.2903/j.efsa.2017.4658
Hart SN, Li Y, Nakamoto K et al (2010) A comparison of whole genome gene expression profiles of HepaRG cells and HepG2 cells to primary human hepatocytes and human liver tissues. Drug Metab Dispos 38:988–994. https://doi.org/10.1124/dmd.109.031831
Hartung T, Tsatsakis AM (2021) The state of the scientific revolution in toxicology. Altex 38:379–386. https://doi.org/10.14573/altex.2106101
Horning JL, Sahoo SK, Vijayaraghavalu S et al (2008) 3-D tumor model for in vitro evaluation of anticancer drugs. Mol Pharm 5:849–862. https://doi.org/10.1021/mp800047v
Josse R, Rogue A, Lorge E, Guillouzo A (2012) An adaptation of the human HepaRG cells to the in vitro micronucleus assay. Mutagenesis 27:295–304. https://doi.org/10.1093/mutage/ger076
Kanebratt KP, Andersson TB (2008) Evaluation of HepaRG cells as an in vitro model for human drug metabolism studies. Drug Metab Dispos 36:1444–1452. https://doi.org/10.1124/dmd.107.020016
Krewski D, Andersen ME, Tyshenko MG et al (2020) Toxicity testing in the 21st century: progress in the past decade and future perspectives. Springer, Berlin
Le Hégarat L, Dumont J, Josse R et al (2010) Assessment of the genotoxic potential of indirect chemical mutagens in HepaRG cells by the comet and the cytokinesis-block micronucleus assays. Mutagenesis 25:555–560. https://doi.org/10.1093/mutage/geq039
Le Hégarat L, Mourot A, Huet S et al (2014) Performance of comet and micronucleus assays in metabolic competent HepaRG cells to predict in vivo genotoxicity. Toxicol Sci 138:300–309. https://doi.org/10.1093/toxsci/kfu004
Leite SB, Wilk-Zasadna I, Zaldivar JM et al (2012) Three-dimensional HepaRG model as an attractive tool for toxicity testing. Toxicol Sci 130:106–116. https://doi.org/10.1093/toxsci/kfs232
Lübberstedt M, Müller-Vieira U, Mayer M et al (2011) HepaRG human hepatic cell line utility as a surrogate for primary human hepatocytes in drug metabolism assessment in vitro. J Pharmacol Toxicol Methods 63:59–68. https://doi.org/10.1016/j.vascn.2010.04.013
Malloy T, Zaunbrecher V, Beryt E et al (2017) Advancing alternatives analysis: the role of predictive toxicology in selecting safer chemical products and processes. Integr Environ Assess Manag 13:915–925. https://doi.org/10.1002/ieam.1923
Mandon M, Huet S, Dubreil E et al (2019) Three-dimensional HepaRG spheroids as a liver model to study human genotoxicity in vitro with the single cell gel electrophoresis assay. Sci Rep 9:10548. https://doi.org/10.1038/s41598-019-47114-7
Mottram DS, Wedzicha BL, Dodson AT (2002) Acrylamide is formed in the Maillard reaction. Nature 419:448–449. https://doi.org/10.1038/419448a
Mueller D, Krämer L, Hoffmann E et al (2014) 3D organotypic HepaRG cultures as in vitro model for acute and repeated dose toxicity studies. Toxicol Vitr 28:104–112. https://doi.org/10.1016/j.tiv.2013.06.024
Ngo LP, Owiti NA, Swartz C et al (2020) Sensitive CometChip assay for screening potentially carcinogenic DNA adducts by trapping DNA repair intermediates. Nucleic Acids Res 48:e13. https://doi.org/10.1093/nar/gkz1077
Oliva-Vilarnau N, Vorrink SU, Ingelman-Sundberg M, Lauschke VM (2020) A 3D cell culture model identifies Wnt/β-catenin mediated inhibition of p53 as a critical step during human hepatocyte regeneration. Adv Sci 7:2000248. https://doi.org/10.1002/advs.202000248
Ott LM, Ramachandran K, Stehno-Bittel L (2017) An Automated multiplexed hepatotoxicity and CYP induction assay using HepaRG cells in 2D and 3D. SLAS Discov Adv Sci Drug Discov 22:614–625. https://doi.org/10.1177/2472555217701058
R Core Team (2021) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/.
Ramaiahgari SC, Waidyanatha S, Dixon D et al (2017) Three-dimensional (3D) HepaRG spheroid model with physiologically relevant xenobiotic metabolism competence and hepatocyte functionality for liver toxicity screening. Toxicol Sci 159:124–136. https://doi.org/10.1093/toxsci/kfx122
Rendic S, Guengerich FP (2012) Contributions of human enzymes in carcinogen metabolism. Chem Res Toxicol 25:1316–1383. https://doi.org/10.1021/tx300132k
Rose S, Cuvellier M, Ezan F et al (2021) DMSO-free highly differentiated HepaRG spheroids for chronic toxicity, liver functions and genotoxicity studies. Arch Toxicol. https://doi.org/10.1007/s00204-021-03178-x
Seo JE, Tryndyak V, Wu Q et al (2019) Quantitative comparison of in vitro genotoxicity between metabolically competent HepaRG cells and HepG2 cells using the high-throughput high-content CometChip assay. Arch Toxicol 93:1433–1448. https://doi.org/10.1007/s00204-019-02406-9
Sevior DK, Pelkonen O, Ahokas JT (2012) Hepatocytes: the powerhouse of biotransformation. Int J Biochem Cell Biol 44:257–261. https://doi.org/10.1016/j.biocel.2011.11.011
Sumner SCJ, Fennell TR, Moore TA et al (1999) Role of cytochrome P450 2E1 in the metabolism of acrylamide and acrylonitrile in mice. Chem Res Toxicol 12:1110–1116. https://doi.org/10.1021/tx990040k
Waldherr M et al (2018) Use of HuH6 and other human-derived hepatoma lines for the detection of genotoxins: a new hope for laboratory animals? Arch Toxicol 92:921–934
Wang Z, Walker GW, Muir DCG, Nagatani-Yoshida K (2020) Toward a global understanding of chemical pollution: a first comprehensive analysis of national and regional chemical inventories. Environ Sci Technol 54:2575–2584. https://doi.org/10.1021/acs.est.9b06379
Wood DK, Weingeist DM, Bhatia SN, Engelward BP (2010) Single cell trapping and DNA damage analysis using microwell arrays. Proc Natl Acad Sci USA 107:10008–10013. https://doi.org/10.1073/pnas.1004056107
Zaunbrecher V, Beryt E, Parodi D et al (2017) Has toxicity testing moved into the 21st century? A survey and analysis of perceptions in the field of toxicology. Environ Health Perspect 125:087024. https://doi.org/10.1289/EHP1435
Acknowledgements
The authors thank Sylvie Huet for technical assistance with HepaRG cell culture.
Funding
This work (“PLATOX3D” project) was funded by a grant (number SAD19003) from the Brittany region (France).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethic approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Barranger, A., Le Hégarat, L. Towards better prediction of xenobiotic genotoxicity: CometChip technology coupled with a 3D model of HepaRG human liver cells. Arch Toxicol 96, 2087–2095 (2022). https://doi.org/10.1007/s00204-022-03292-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00204-022-03292-4