Abstract
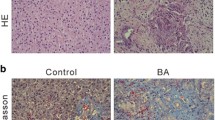
Liver fibrosis is the final common pathway for almost all causes of chronic liver injury. This chronic disease is characterized by excessive deposition of extracellular matrix components mainly due to transdifferentiation of quiescent hepatic stellate cell into myofibroblasts-like cells, which in turn is driven by cell death and inflammation. In the last few years, paracrine signaling through pannexin1 channels has emerged as a key player in the latter processes. The current study was set up to investigate the role of pannexin1 signaling in liver fibrosis. Wild-type and whole body pannexin1 knock-out mice were treated with carbon tetrachloride or subjected to bile duct ligation. Evaluation of the effects of pannexin1 deletion was based on a number of clinically relevant read-outs, including markers of liver damage, histopathological analysis, oxidative stress, inflammation and regenerative capacity. In parallel, to elucidate the molecular pathways affected by pannexin1 deletion as well as to mechanistically anchor the clinical observations, whole transcriptome analysis of liver tissue was performed. While pannexin1 knock-out mice treated with carbon tetrachloride displayed reduced collagen content, hepatic stellate cell activation, inflammation and hepatic regeneration, bile duct ligated counterparts showed increased hepatocellular injury and antioxidant enzyme activity with a predominant immune response. Gene expression profiling revealed a downregulation of fibrotic and immune responses in pannexin1 knock-out mice treated with carbon tetrachloride, whereas bile duct ligated pannexin1-deficient animals showed a pronounced inflammatory profile. This study shows for the first time an etiology-dependent role for pannexin1 signaling in experimental liver fibrosis.














Similar content being viewed by others
Abbreviations
- α-SMA:
-
Alpha smooth muscle actin
- ALT:
-
Alanine aminotransferase
- ALP:
-
Alkaline phosphatase
- ANOVA:
-
Analysis of variance
- ASC:
-
Apoptosis-associated speck-like protein containing a C-terminal caspase-recruitment domain
- AST:
-
Aspartate aminotransferase
- ATP:
-
Adenosine-5′-triphosphate
- BDL:
-
Bile duct ligation
- Casp1:
-
Caspase 1
- CCl4 :
-
Carbon tetrachloride
- Clec7a:
-
Dectin-1
- Col1a1:
-
Collagen type 1 alpha 1
- Cxcl14:
-
Chemokine ligand 14
- GPx:
-
Glutathione peroxidase
- GR:
-
Glutathione reductase
- HSCs:
-
Hepatic stellate cells
- IL:
-
Interleukin
- ip:
-
Intraperitoneally
- Lbp:
-
Lipopolysaccharide-binding protein
- Loxl2:
-
Lysyl oxidase-like 2
- Ly96:
-
Lymphocyte antigen 96
- MCP-1:
-
Monocyte chemoattractant protein 1
- MIP1γ:
-
Macrophage protein 1 gamma
- Nalp3:
-
NACHT, LRR, and pyrin domain-containing protein 3
- n :
-
Number or repeats
- NF-κB:
-
Nuclear factor kappa B
- Panx:
-
Pannexin
- p :
-
Probability
- RT-qPCR:
-
Reverse transcription quantitative real-time polymerase chain reaction
- S100a9:
-
S100 calcium-binding protein A9
- SEM:
-
Standard error of mean
- SOD:
-
Superoxide dismutase
- sTNF-RI/-RII:
-
Soluble TNF receptor 1/2
- TCA-3:
-
T-cell activation protein 3
- TECK:
-
Thymus-expressed chemokine
- TLR:
-
Toll-like receptor
- TNFα:
-
Tumor necrosis factor alpha
- WT:
-
Wild-type
References
Bao Y, Chen Y, Ledderose C, Li L, Junger WG (2013) Pannexin 1 channels link chemoattractant receptor signaling to local excitation and global inhibition responses at the front and back of polarized neutrophils. J Biol Chem 288:22650–22657. https://doi.org/10.1074/jbc.M113.476283
Cai SY, Ouyang X, Chen Y et al (2017) Bile acids initiate cholestatic liver injury by triggering a hepatocyte-specific inflammatory response. JCI Insight 2:e90780. https://doi.org/10.1172/jci.insight.90780
Canbay A, Taimr P, Torok N, Higuchi H, Friedman S, Gores GJ (2003) Apoptotic body engulfment by a human stellate cell line is profibrogenic. Lab Invest 83:655–663
Chekeni FB, Elliott MR, Sandilos JK et al (2010) Pannexin 1 channels mediate ‘find-me’ signal release and membrane permeability during apoptosis. Nature 467:863–867. https://doi.org/10.1038/nature09413
Chen W, Xu WH (2015) β-Actin as a loading control: less than 2 µg of total protein should be loaded. Electrophoresis 36:2046–2049. https://doi.org/10.1002/elps.201500138
Cogliati B, Da Silva TC, Aloia TP et al (2011) Morphological and molecular pathology of CCL4-induced hepatic fibrosis in connexin43-deficient mice. Microsc Res Tech 74:421–429. https://doi.org/10.1002/jemt.20926
Cogliati B, Crespo Yanguas S, Da Silva TC et al (2016) Connexin32 deficiency exacerbates carbon tetrachloride-induced hepatocellular injury and liver fibrosis in mice. Toxicol Mech Methods 26:362–370. https://doi.org/10.1080/15376516.2016.1190991
Crespo Yanguas S, Willebrords J, Johnstone SR et al (2017) Pannexin1 as mediator of inflammation and cell death. Biochim Biophys Acta 1864:51–61. https://doi.org/10.1016/j.bbamcr.2016.10.006
Csak T, Ganz M, Pespisa J, Kodys K, Dolganiuc A, Szabo G (2011) Fatty acid and endotoxin activate inflammasomes in mouse hepatocytes that release danger signals to stimulate immune cells. Hepatology 54:133–144. https://doi.org/10.1002/hep.24341
Dahl G (2015) ATP release through pannexon channels. Philos Trans R Soc Lond B Biol Sci 370:20140191. https://doi.org/10.1098/rstb.2014.0191
Davalos D, Grutzendler J, Yang G et al (2005) ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci 8:752–758
Dolmatova E, Spagnol G, Boassa D et al (2012) Cardiomyocyte ATP release through pannexin 1 aids in early fibroblast activation. Am J Physiol Heart Circ Physiol 303:H1208–H1218. https://doi.org/10.1152/ajpheart.00251.2012
Dunning S, Ur Rehman A, Tiebosch MH et al (2013) Glutathione and antioxidant enzymes serve complementary roles in protecting activated hepatic stellate cells against hydrogen peroxide-induced cell death. Biochim Biophys Acta 1832:2027–2034. https://doi.org/10.1016/j.bbadis.2013.07.008
Dvoriantchikova G, Ivanov D, Barakat D et al (2012) Genetic ablation of Pannexin1 protects retinal neurons from ischemic injury. PLoS One 7:e31991. https://doi.org/10.1371/journal.pone.0031991
Eaton SL, Roche SL, Llavero Hurtado M et al (2013) Total protein analysis as a reliable loading control for quantitative fluorescent Western blotting. PLoS One 8:e72457. https://doi.org/10.1371/journal.pone.0072457
Gandhi CR (2012) Oxidative stress and hepatic stellate cells: a paradoxical relationship. Trends Cell Mol Biol 7:1–10
Hao H, Cao L, Jiang C et al (2017) Farnesoid X receptor regulation of the NLRP3 inflammasome underlies cholestasis-associated sepsis. Cell Metab 25:856–867. https://doi.org/10.1016/j.cmet.2017.03.007
Hara T, Bacon KB, Cho LC et al (1995) Molecular cloning and functional characterization of a novel member of the C-C chemokine family. J Immunol 155:5352–5358
Imamura M, Ogawa T, Sasaguri Y, Chayama K, Ueno H (2005) Suppression of macrophage infiltration inhibits activation of hepatic stellate cells and liver fibrogenesis in rats. Gastroenterology 128:138–146. https://doi.org/10.1053/j.gastro.2004.10.005
Jiang S, Zhang Y, Zheng JH et al (2017) Potentiation of hepatic stellate cell activation by extracellular ATP is dependent on P2 × 7R-mediated NLRP3 inflammasome activation. Pharmacol Res 117:82–93. https://doi.org/10.1016/j.phrs.2016.11.040
Karlmark KR, Weiskirchen R, Zimmermann HW et al (2009) Hepatic recruitment of the inflammatory Gr1 + monocyte subset upon liver injury promotes hepatic fibrosis. Hepatology 50:261–274. https://doi.org/10.1002/hep.22950
Kim HY, Kim SJ, Lee SM (2015) Activation of NLRP3 and AIM2 inflammasomes in Kupffer cells in hepatic ischemia/reperfusion. FEBS J 282:259–270. https://doi.org/10.1111/febs.13123
Kowal JM, Haanes KA, Christensen NM, Novak I (2015) Bile acid effects are mediated by ATP release and purinergic signalling in exocrine pancreatic cells. Cell Commun Signal 13:28. https://doi.org/10.1186/s12964-015-0107-9
Krenkel O, Tacke F (2017) Liver macrophages in tissue homeostasis and disease. Nat Rev Immunol 17:306–321. https://doi.org/10.1038/nri.2017.11
Krick S, Wang J, St-Pierre M, Gonzalez C, Dahl G, Salathe M (2016) Dual oxidase 2 (Duox2) regulates pannexin 1-mediated ATP release in primary human airway epithelial cells via changes in intracellular pH and not H2O2 production. J Biol Chem 291:6423–6432. https://doi.org/10.1074/jbc.M115.664854
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Maes M, McGill MR, da Silva TC et al (2016) Involvement of connexin43 in acetaminophen-induced liver injury. Biochim Biophys Acta 1862:1111–1121. https://doi.org/10.1016/j.bbadis.2016.02.007
Maes M, McGill MR, da Silva TC et al (2017) Inhibition of pannexin1 channels alleviates acetaminophen-induced hepatotoxicity. Arch Toxicol 91:2245–2261. https://doi.org/10.1007/s00204-016-1885-6
Moles A, Murphy L, Wilson CL et al (2014) A TLR2/S100A9/CXCL-2 siganling for neutrophil recruitment in acute and chronic liver injury in the mouse. J Hepatol 60:782–791. https://doi.org/10.1016/j.jhep.2013.12.005
Pradere JP, Kluwe J, De Minicis S et al (2013) Hepatic macrophages but not dendritic cells contribute to liver fibrosis by promoting the survival of activated hepatic stellate cells in mice. Hepatology 58:1461–1473. https://doi.org/10.1002/hep.26429
Puche JE, Lee YA, Jiao J et al (2013) A novel murine model to deplete hepatic stellate cells uncovers their role in amplifying liver damage in mice. Hepatology 57:339–350. https://doi.org/10.1002/hep.26053
Rivero-Gutiérrez B, Anzola A, Martínez-Augustin O, de Medina FS (2014) Stain-free detection as loading control alternative to Ponceau and housekeeping protein immunodetection in Western blotting. Anal Biochem 467:1–3. https://doi.org/10.1016/j.ab.2014.08.027
Saito JM, Bostick MK, Campe CB, XU J, Maher JJ (2003) Infiltrating neutrophils in bile duct-ligated livers do not promote hepatic fibrosis. Hepatol Res 25:180–191. https://doi.org/10.1016/S1386-6346(02)00247-4
Schindelin J, Rueden CT, Hiner MC, Eliceiri KW (2015) The ImageJ ecosystem: an open platform for biomedical image analysis. Mol Reprod Dev 82:518–529. https://doi.org/10.1002/mrd.22489
Seifert L, Deutsch M, Alothman S et al (2015) Dectin-1 regulates hepatic fibrosis and hepatocarcinogenesis by suppressing TLR4 signaling pathways. Cell Rep 13:1909–1921. https://doi.org/10.1016/j.celrep.2015.10.058
Seki E, Schwabe RF (2015) Hepatic inflammation and fibrosis: functional links and key pathways. Hepatology 61:1066–1079. https://doi.org/10.1002/hep.27332
Simard JC, Cesaro A, Chapeton-Montes J et al (2013) S100A8 and S100A9 induce cytokine expression and regulate the NLRP3 inflammasome via ROS-dependent activation of NF-κB. PLoS One 8:e72138. https://doi.org/10.1371/journal.pone.0072138
Tag CG, Sauer-Lehnen S, Weiskirchen S et al (2015) Bile duct ligation in mice: induction of inflammatory liver injury and fibrosis by obstructive cholestasis. J Vis Exp 96:52438. https://doi.org/10.3791/52438
Taylor SC, Posch A (2014) The design of a quantitative western blot experiment. Biomed Res Int 2014:361590. https://doi.org/10.1155/2014/361590
Taylor SC, Berkelman T, Yadav G, Hammond M (2013) A defined methodology for reliable quantification of Western blot data. Mol Biotechnol 55:217–226. https://doi.org/10.1007/s12033-013-9672-6
Willebrords J, Cogliati B, Pereira IVA et al (2017a) Inhibition of connexin hemichannels alleviates non-alcoholic steatohepatitis in mice. Sci Rep 7:8268. https://doi.org/10.1038/s41598-017-08583-w
Willebrords J, Maes M, Pereira IVA et al (2017b) Protective effect of genetic deletion of pannexin1 in experimental mouse models of acute and chronic liver disease. Biochim Biophys Acta 1864:819–830. https://doi.org/10.1016/j.bbadis.2017.12.013
Woolbright BL, Jaeschke H (2012) Novel insight into mechanisms of cholestatic liver injury. World J Gastroenterol 18:4985–4993. https://doi.org/10.3748/wjg.v18.i36.4985
Wree A, Eguchi A, McGeough MD et al (2014) NLRP3 inflammasome activation results in hepatocyte pyroptosis, liver inflammation, and fibrosis in mice. Hepatology 59:898–910. https://doi.org/10.1002/hep.26592
Xiao F, Waldrop SL, Khimji AK, Kilic G (2012) Pannexin1 contributes to pathophysiological ATP release in lipoapoptosis induced by saturated free fatty acids in liver cells. Am J Physiol Cell Physiol 303:C1034–C1044. https://doi.org/10.1152/ajpcell.00175.2012
Xiao F, Waldrop SL, Bronk SF, Gores GJ, Davis LS, Kilic G (2015) Lipoapoptosis induced by saturated free fatty acids stimulates monocyte migration: a novel role for Pannexin1 in liver cells. Purinergic Signal 11:347–359. https://doi.org/10.1007/s11302-015-9456-5
Xie G, Wang X, Wang L et al (2012) Role of differentiation of liver sinusoidal endothelial cells in progression and regression of hepatic fibrosis in rats. Gastroenterology 142:918–927.e6. https://doi.org/10.1053/j.gastro.2011.12.017
Yan HH, Jiang J, Pang Y et al (2015) CCL9 induced by TGF-β signaling in myeloid cells enhances tumor cell survival in the premetastatic organ. Cancer Res 75:5283–5298. https://doi.org/10.1158/0008-5472.CAN-15-2282-T
Acknowledgements
This work was supported by the grants of the “Fundação de Auxílio à Pesquisa do Estado de São Paulo” (FAPESP Grants 14/23890-4; 14/23887-3 and SPEC 13/50420-6), the European Research Council (ERC Starting Grant 335476), the Fund for Scientific Research-Flanders (FWO Grants G009514N and G010214N) and the University Hospital of the Vrije Universiteit Brussel-Belgium (“Willy Gepts Fonds” UZ-VUB). The authors thank Miss Tineke Vanhalewyn, Miss Dinja De Win and Mr Steven Branson for their dedicated technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Electronic supplementary material
Below is the link to the electronic supplementary material.
204_2018_2255_MOESM1_ESM.pdf
Western blots from p-NF-κBp65 S536 protein expression in liver fibrosis. WT and Panx1-/- mice (n = 10-15) were administered 20% CCl4 ip at a gradually increased dose for 8 weeks or subjected to BDL for 20 days. Representative blot membranes expressing p-NF-κBp65 S536 proteins, total protein loading and β-actin proteins from a the CCl4 model and b the BDL model (PDF 230 KB)
204_2018_2255_MOESM3_ESM.docx
Gene modulation by the CCl4 treatment independent of background: comparison between WT CCl4 vs WT-oil and Panx1-/- CCl4 vs WT-oil (DOCX 25 KB)
204_2018_2255_MOESM5_ESM.docx
Gene modulation by the BDL procedure independent of background: comparison between WT BDL vs WT-sham and Panx1-/- BDL vs WT-sham (DOCX 45 KB)
Rights and permissions
About this article
Cite this article
Crespo Yanguas, S., da Silva, T.C., Pereira, I.V.A. et al. Genetic ablation of pannexin1 counteracts liver fibrosis in a chemical, but not in a surgical mouse model. Arch Toxicol 92, 2607–2627 (2018). https://doi.org/10.1007/s00204-018-2255-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00204-018-2255-3