Abstract
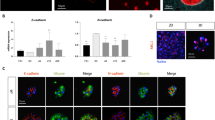
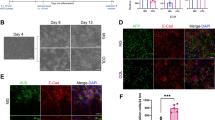
Human primary hepatocytes represent a gold standard in in vitro liver research. Due to their low availability and high costs alternative liver cell models with comparable morphological and biochemical characteristics have come into focus. The human hepatocarcinoma cell line HepG2 is often used as a liver model for toxicity studies. However, under two-dimensional (2D) cultivation conditions the expression of xenobiotic-metabolizing enzymes and typical liver markers such as albumin is very low. Cultivation for 21 days in a three-dimensional (3D) Matrigel culture system has been reported to strongly increase the metabolic competence of HepG2 cells. In our present study we further compared HepG2 cell cultivation in three different 3D systems: collagen, Matrigel and Alvetex culture. Cell morphology, albumin secretion, cytochrome P450 monooxygenase enzyme activities, as well as gene expression of xenobiotic-metabolizing and liver-specific enzymes were analyzed after 3, 7, 14, and 21 days of cultivation. Our results show that the previously reported increase of metabolic competence of HepG2 cells is not primarily the result of 3D culture but a consequence of the duration of cultivation. HepG2 cells grown for 21 days in 2D monolayer exhibit comparable biochemical characteristics, CYP activities and gene expression patterns as all 3D culture systems used in our study. However, CYP activities did not reach the level of HepaRG cells. In conclusion, the increase of metabolic competence of the hepatocarcinoma cell line HepG2 is not due to 3D cultivation but rather a result of prolonged cultivation time.






Similar content being viewed by others
Abbreviations
- 2D:
-
Two-dimensional
- 3D:
-
Three-dimensional
- ACTB:
-
β-Actin
- AhR:
-
Aryl hydrocarbon receptor
- Alv:
-
Alvetex
- CAR:
-
Constitutive androstane receptor
- Col:
-
Collagen
- CTB:
-
CellTiter-Blue
- CYP:
-
Cytochrome-P450 monooxygenase(s)
- DPPIV:
-
Dipeptidyl-peptidase IV
- GAPDH:
-
Glyceraldehyde 3-phosphate dehydrogenase
- Mg:
-
Matrigel
- OAZ1:
-
Ornithine decarboxylase antizyme 1
- PXR:
-
Pregnane-X receptor
- qPCR:
-
Real-time quantitative PCR
- RFU:
-
Relative fluorescence units
- SD:
-
Standard deviation
- UGT:
-
UDP-glucuronosyltransferase(s)
- ZO-1:
-
Zona occludens-1
References
Aninat C, Piton A, Glaise D et al (2006) Expression of cytochromes P450, conjugating enzymes and nuclear receptors in human hepatoma HepaRG cells. Drug Metab Dispos 34(1):75–83. doi:10.1124/dmd.105.006759
Bissell DM, Arenson DM, Maher JJ, Roll FJ (1987) Support of cultured hepatocytes by a laminin-rich gel. Evidence for a functionally significant subendothelial matrix in normal rat liver. J Clin Invest 79(3):801–812. doi:10.1172/jci112887
Bissell MJ, Rizki A, Mian IS (2003) Tissue architecture: the ultimate regulator of breast epithelial function. Curr Opin Cell Biol 15(6):753–762
Breslin S, O’Driscoll L (2013) Three-dimensional cell culture: the missing link in drug discovery. Drug Discov Today 18(5–6):240–249. doi:10.1016/j.drudis.2012.10.003
Clayton DF, Darnell JE Jr (1983) Changes in liver-specific compared to common gene transcription during primary culture of mouse hepatocytes. Mol Cell Biol 3(9):1552–1561
Clayton DF, Weiss M, Darnell JE Jr (1985) Liver-specific RNA metabolism in hepatoma cells: variations in transcription rates and mRNA levels. Mol Cell Biol 5(10):2633–2641
Daus AW, Goldhammer M, Layer PG, Thielemann C (2011) Electromagnetic exposure of scaffold-free three-dimensional cell culture systems. Bioelectromagnetics 32(5):351–359. doi:10.1002/bem.20649
Debnath J, Brugge JS (2005) Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer 5(9):675–688. doi:10.1038/nrc1695
Donato MT, Lahoz A, Castell JV, Gomez-Lechon MJ (2008) Cell lines: a tool for in vitro drug metabolism studies. Curr Drug Metab 9(1):1–11
Feidt DM, Klein K, Hofmann U et al (2010) Profiling induction of cytochrome p450 enzyme activity by statins using a new liquid chromatography-tandem mass spectrometry cocktail assay in human hepatocytes. Drug Metab Dispos 38(9):1589–1597. doi:10.1124/dmd.110.033886
Godoy P, Hengstler JG, Ilkavets I et al (2009) Extracellular matrix modulates sensitivity of hepatocytes to fibroblastoid dedifferentiation and transforming growth factor beta-induced apoptosis. Hepatology 49(6):2031–2043. doi:10.1002/hep.22880
Godoy P, Hewitt N, Albrecht U et al (2013) Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol 87(8):1315–1530. doi:10.1007/s00204-013-1078-5
Gomez-Lechon MJ, Donato MT, Castell JV, Jover R (2003) Human hepatocytes as a tool for studying toxicity and drug metabolism. Curr Drug Metab 4(4):292–312
Guguen-Guillouzo C, Guillouzo A (2010) General review on in vitro hepatocyte models and their applications. Methods Mol Biol 640:1–40. doi:10.1007/978-1-60761-688-7_1
Hammad S, Hoehme S, Friebel A et al (2014) Protocols for staining of bile canalicular and sinusoidal networks of human, mouse and pig livers, three-dimensional reconstruction and quantification of tissue microarchitecture by image processing and analysis. Arch Toxicol 88(5):1161–1183. doi:10.1007/s00204-014-1243-5
Harma V, Virtanen J, Makela R et al (2010) A comprehensive panel of three-dimensional models for studies of prostate cancer growth, invasion and drug responses. PLoS ONE 5(5):e10431. doi:10.1371/journal.pone.0010431
Hughes CS, Postovit LM, Lajoie GA (2010) Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics 10(9):1886–1890. doi:10.1002/pmic.200900758
Ivascu A, Kubbies M (2006) Rapid generation of single-tumor spheroids for high-throughput cell function and toxicity analysis. J Biomol Screen 11(8):922–932. doi:10.1177/1087057106292763
Kanebratt KP, Andersson TB (2008) Evaluation of HepaRG cells as an in vitro model for human drug metabolism studies. Drug Metab Dispos 36(7):1444–1452. doi:10.1124/dmd.107.020016
Klein K, Zanger UM (2013) Pharmacogenomics of cytochrome P450 3A4: recent progress toward the “missing heritability” problem. Front Genet 4:12. doi:10.3389/fgene.2013.00012
Knight E, Murray B, Carnachan R, Przyborski S (2011) Alvetex(R): polystyrene scaffold technology for routine three dimensional cell culture. Methods Mol Biol 695:323–340. doi:10.1007/978-1-60761-984-0_20
LeCluyse EL (2001) Human hepatocyte culture systems for the in vitro evaluation of cytochrome P450 expression and regulation. Eur J Pharm Sci 13(4):343–368
Lin RZ, Chou LF, Chien CC, Chang HY (2006) Dynamic analysis of hepatoma spheroid formation: roles of E-cadherin and beta1-integrin. Cell Tissue Res 324(3):411–422. doi:10.1007/s00441-005-0148-2
Loessner D, Stok KS, Lutolf MP, Hutmacher DW, Clements JA, Rizzi SC (2010) Bioengineered 3D platform to explore cell-ECM interactions and drug resistance of epithelial ovarian cancer cells. Biomaterials 31(32):8494–8506. doi:10.1016/j.biomaterials.2010.07.064
McClelland R, Wauthier E, Uronis J, Reid L (2008) Gradients in the liver’s extracellular matrix chemistry from periportal to pericentral zones: influence on human hepatic progenitors. Tissue Eng Part A 14(1):59–70. doi:10.1089/ten.a.2007.0058
Mowbray CA (2011) In vitro systems to predict hepatotoxicity: models based on hepatocarcinoma cell lines. Newcastle University, Faculty of Medical Sciences, Institute of Cell and Molecular Biosciences, https://theses.ncl.ac.uk/dspace/bitstream/10443/1337/1/Mowbray%2011.pdf
Ramaiahgari SC, den Braver MW, Herpers B et al (2014) A 3D in vitro model of differentiated HepG2 cell spheroids with improved liver-like properties for repeated dose high-throughput toxicity studies. Arch Toxicol 88(5):1083–1095. doi:10.1007/s00204-014-1215-9
Ravi M, Paramesh V, Kaviya SR, Anuradha E, Solomon FD (2015) 3D cell culture systems: advantages and applications. J Cell Physiol 230(1):16–26. doi:10.1002/jcp.24683
Weaver VM, Petersen OW, Wang F et al (1997) Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol 137(1):231–245
Wilkening S, Bader A (2003) Influence of culture time on the expression of drug-metabolizing enzymes in primary human hepatocytes and hepatoma cell line HepG2. J Biochem Mol Toxicol 17(4):207–213. doi:10.1002/jbt.10085
Acknowledgments
This work was financed by the Federal Institute for Risk Assessment (grant number 1322-591) and by the Robert Bosch Foundation, Stuttgart. We thank Beatrice Rosskopp, Markus König (Stuttgart) and Igor Liebermann (Stuttgart) for their excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Luckert, C., Schulz, C., Lehmann, N. et al. Comparative analysis of 3D culture methods on human HepG2 cells. Arch Toxicol 91, 393–406 (2017). https://doi.org/10.1007/s00204-016-1677-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00204-016-1677-z