Abstract
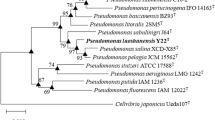
An aerobic, Gram-negative, non-sporulating, motile, rod-shaped and lignin-degrading bacterial strain, Pseudomonas sp. CCA1, was isolated from leaf soil collected in Japan. This strain grew at 20–45 °C (optimum 20 °C), at pH 5.0–10.0 (optimum pH 5.0), and in the presence of 2% NaCl. Its major cellular fatty acids were C16:0 and summed feature 8 (C18:1ω6c and/or C18:1ω7c). The predominant quinone system was ubiquinone-9. Comparative 16S rRNA gene sequence analysis showed that Pseudomonas sp. CCA1 was related most closely to P. citronellolis NBRC 103043T (98.9%), but multilocus sequence analysis based on fragments of the atpD, gyrA, gyrB and rpoB gene sequences showed strain CCA1 to branch separately from its most closely related Pseudomonas type strains. DNA–DNA hybridization values between Pseudomonas sp. CCA1 and type strains of closely related Pseudomonas species were less than 53%. Based on its phenotypic, chemotaxonomic and phylogenetic features, we propose that Pseudomonas sp. CCA1 represents a novel species within the genus Pseudomonas, for which the name Pseudomonas humi sp. nov. is proposed. The type strain is CCA1 (= HUT 8136T = TBRC 8616T).

Similar content being viewed by others
References
Ahmad M, Taylor CR, Pink D, Burton K, Eastwood D, Bending GD, Bugg TD (2010) Development of novel assays for lignin degradation: comparative analysis of bacterial and fungal lignin degraders. Mol Biosyst 6:815–821
Akita H, Kimura Z, Yusoff MZ, Hoshino T (2016a) Isolation of Pseudomonas sp. strain CCA1 from leaf soil and preliminary characterization its ligninolytic activity. JSM Biotechnol Bioeng 3:1062
Akita H, Kimura Z, Hoshino T (2016b) Draft genome sequence of Pseudomonas sp. strain CCA1, isolated from leaf soil. Genome Announc 4:e01371–16
Arantes V, Jellison J, Goodell B (2012) Peculiarities of brown-rot fungi and biochemical Fenton reaction with regard to their potential as a model for bioprocessing biomass. Appl Microbiol Biotechnol 94:323–338
Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37:911–917
Bugg TD, Ahmad M, Hardiman EM, Singh R (2011) The emerging role for bacteria in lignin degradation and bio-product formation. Curr Opin Biotechnol 22:394–400
Dashtban M, Schraft H, Syed TA, Qin W (2010) Fungal biodegradation and enzymatic modification of lignin. Int J Biochem Mol Biol 1:36–50
Gordon RE, Barnett DA, Handerhan JE, Pang CHN (1974) Nocardia coeliaca, Nocardia autotrophica, and the nocardin strain. Int J Syst Bacteriol 24:54–63
Gupta SK, Kumari R, Prakash O, Lal R (2008) Pseudomonas panipatensis sp. nov., isolated from an oil-contaminated site. Int J Syst Evol Microbiol 58:1339–1345
Hugh R, Leifson E (1964) The proposed neotype strains of Pseudomonas aeruginosa (Schroeter 1872) Migula 1900. Int J Syst Evol Microbiol l4:69–84
Iizuka H, Komagata K (1964) Microbiological studies on petroleum and natural gas. I. Determination of hydrocarbon-utilizing bacteria. J Gen Appl Microbiol 10:207–221
Jones MPA, McCarthy AJ, Cross T (1979) Taxonomic and serological studies on Micropolyspora faeni and Micropolyspora strains from soil bearing the specific epithet rectivirgula. J Gen Microbiol 115:343–354
Kubota M, Kawahara K, Sekiya K, Uchida T, Hattori Y, Futamata H, Hiraishi A (2005) Nocardioides aromaticivorans sp. nov., a dibenzofuran-degrading bacterium isolated from dioxinpolluted environments. Syst Appl Microbiol 28:165–174
Kwon SW, Kim JS, Park IC, Yoon SH, Park DH, Lim CK, Go SJ (2003) Pseudomonas koreensis sp. nov., Pseudomonas umsongensis sp. nov. and Pseudomonas jinjuensis sp. nov., novel species from farm soils in Korea. Int J Syst Evol Microbiol 53:21–27
Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG (1998) Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA 95:3140–3145
Prakash O, Kumari K, Lal R (2007) Pseudomonas delhiensis sp. nov., from a fly ash dumping site of a thermal power plant. Int J Syst Evol Microbiol 57:527–531
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Seubert W (1960) Degradation of isoprenoid compounds by micro-organisms. I. Isolation and characterization of an isoprenoid-degrading bacterium, Pseudomonas citronellolis n. sp. J Bacteriol 79:426–434
Spencer DF, Schnare MN, Gray MW (1984) Pronounced structural similarities between the small subunit ribosomal RNA genes of wheat mitochondria and Escherichia coli. Proc Natl Acad Sci USA 81:493–497
Stolz A, Busse HJ, Kämpfer P (2007) Pseudomonas knackmussii sp. nov. Int J Syst Evol Microbiol 57:572–576
Tamaoka J, Katayama-Fujimura Y, Kuraishi H (1983) Analysis of bacterial menaquinone mixtures by high performance liquid chromatography. J appl Bacteriol 54:31–36
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tian JH, Pourcher AM, Peu P (2016) Isolation of bacterial strains able to metabolize lignin and lignin-related compounds. Lett Appl Microbiol 63:30–37
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Truper HG (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Evol Microbiol 37:463–464
Funding
This work was supported in part by the Science and Technology Research Partnership for Sustainable Development (SATREPS), under the Japan Science and Technology Agency (JST) and Japan International Cooperation Agency (JICA).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by Erko Stackebrandt.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Akita, H., Kimura, Zi. & Hoshino, T. Pseudomonas humi sp. nov., isolated from leaf soil. Arch Microbiol 201, 245–251 (2019). https://doi.org/10.1007/s00203-018-1588-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-018-1588-x